
Overview of Alba v5
Alba v5 is a laser scanning microscope that incorporates several measurement modalities for experimental quantitative biology and material science applications requiring single molecule detection sensitivity. Capable of acquisition from the violet to the near infrared region, it features two independent laser entry ports. Alba v5 is powered by VistaVision, the comprehensive software package for instrument control, image acquisition and processing.
Key Features of Alba v5
Multimodality imaging (Intensity, lifetime and SHG)
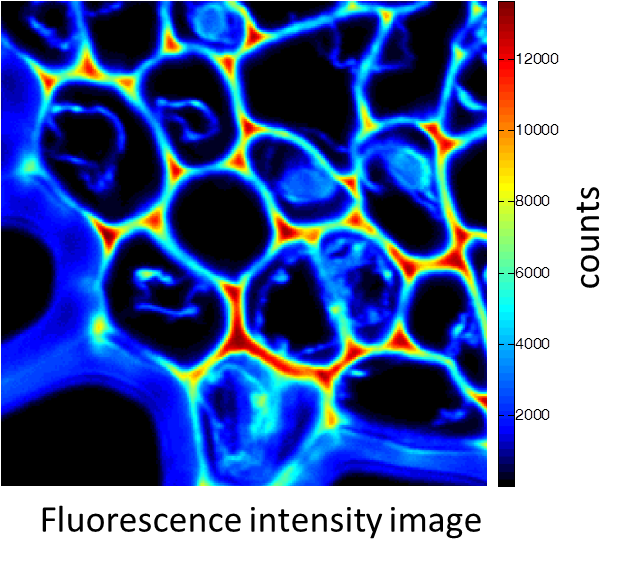
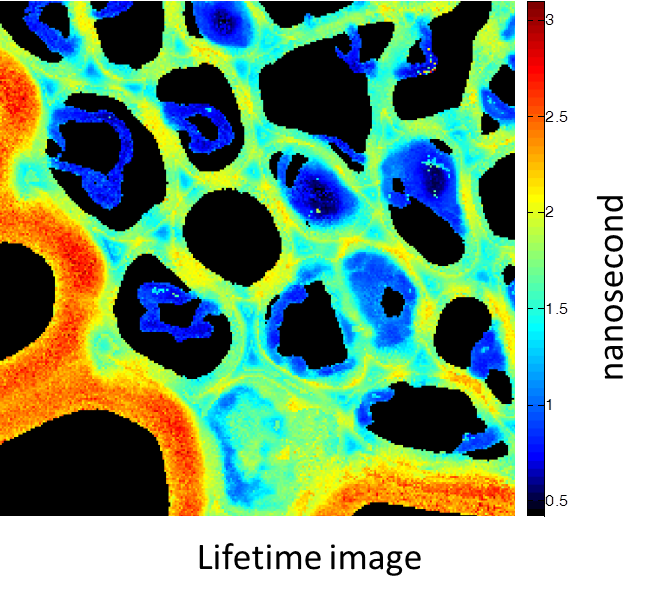
Rhizome cross-section of Convallaria (Lily of the Valley). Images were taken simultaneously using an Alba equipped with a multiphoton laser emitting at 800 nm. The standard fluorescence intensity image (first), the fluorescence lifetime image (second) and the second-harmonic generation (SHG) image (third). All images are 256 × 256 pixels, 40 × 40 µm2.
(courtesy of Dr. Zhang, Beckman Institute, Urbana, IL)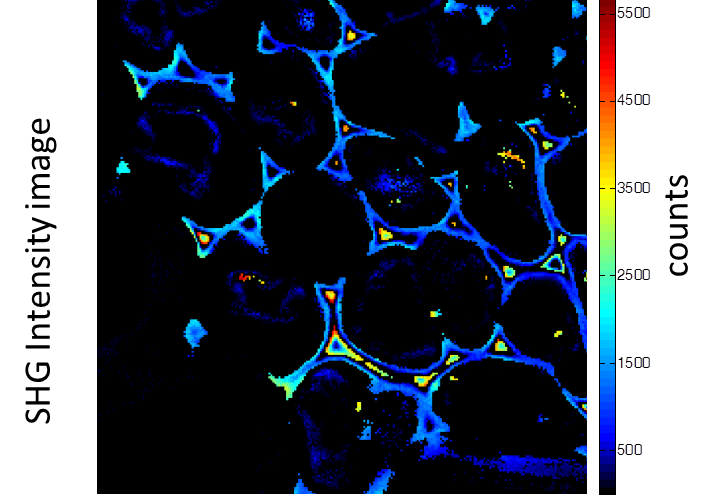
Tissue Imaging
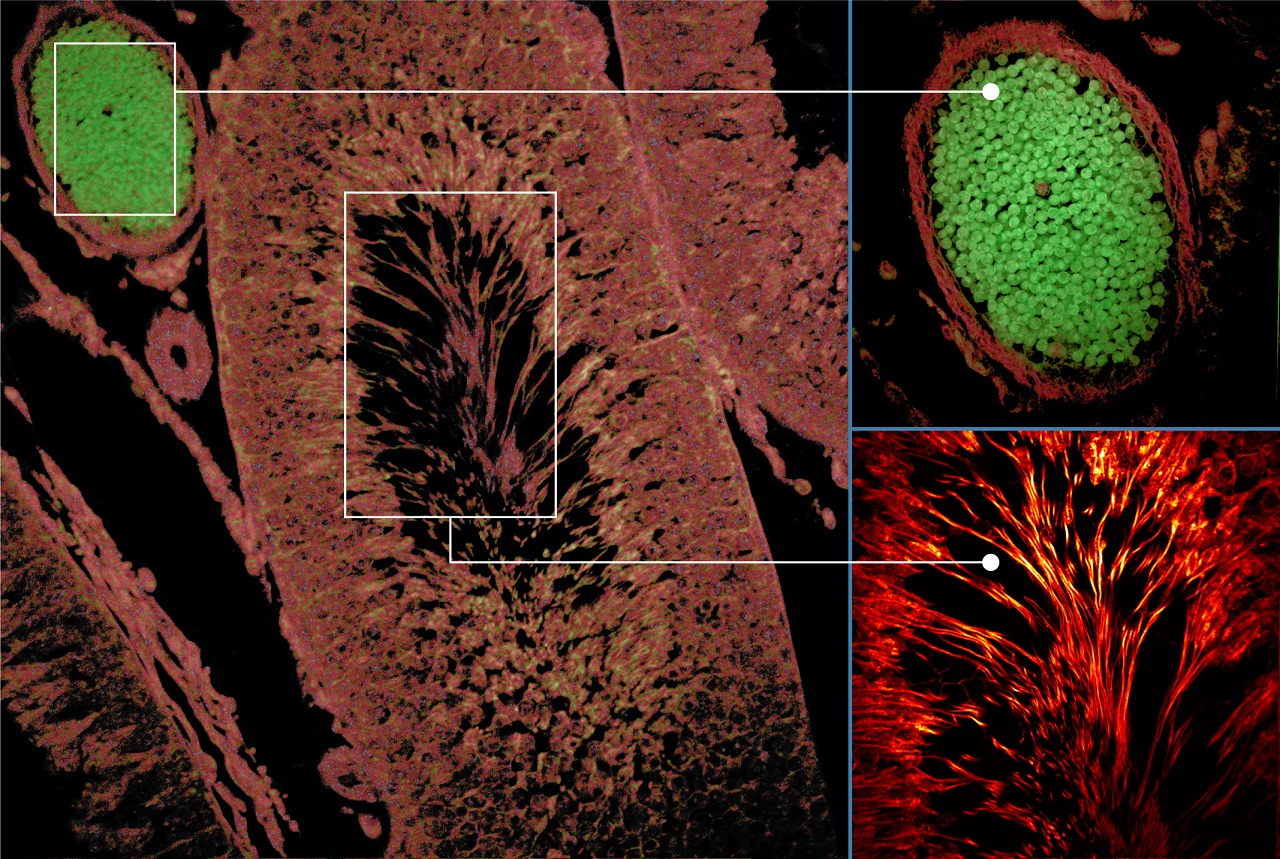
Autofluorescence tissue image: Excitation: 375 nm, Emission: 425 nm, Objective: 20X NA0.75
(courtesy of Dr. Aneesh Alex, GSK)
FLIM-FRET in live cells
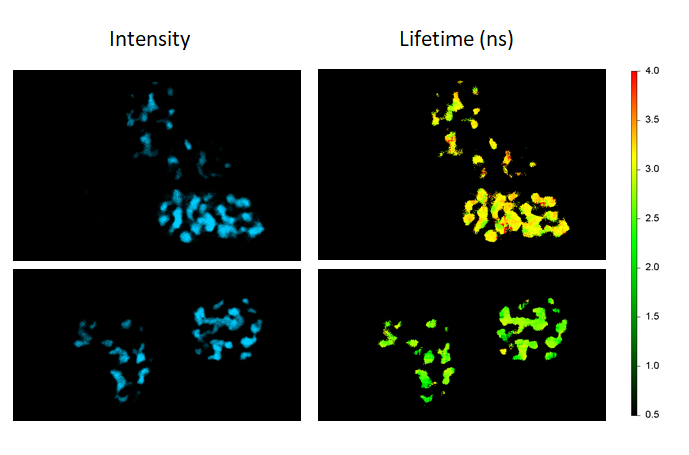
FLIM-FRET maps the FRET efficiencies of Cebp/α proteins co-expressing Cerulean and Venus in live cells to localize their dimerization in the cell nuclei (donor-only control: cells expressing Cebp/α-Cerulean only; FRET sample: cells co-expressing Cebp/α-Cerulean and Cebp/α-Venus).
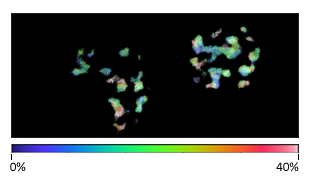
Multiplex imaging by Time-Resolved Unmixing using the Phasor Plot
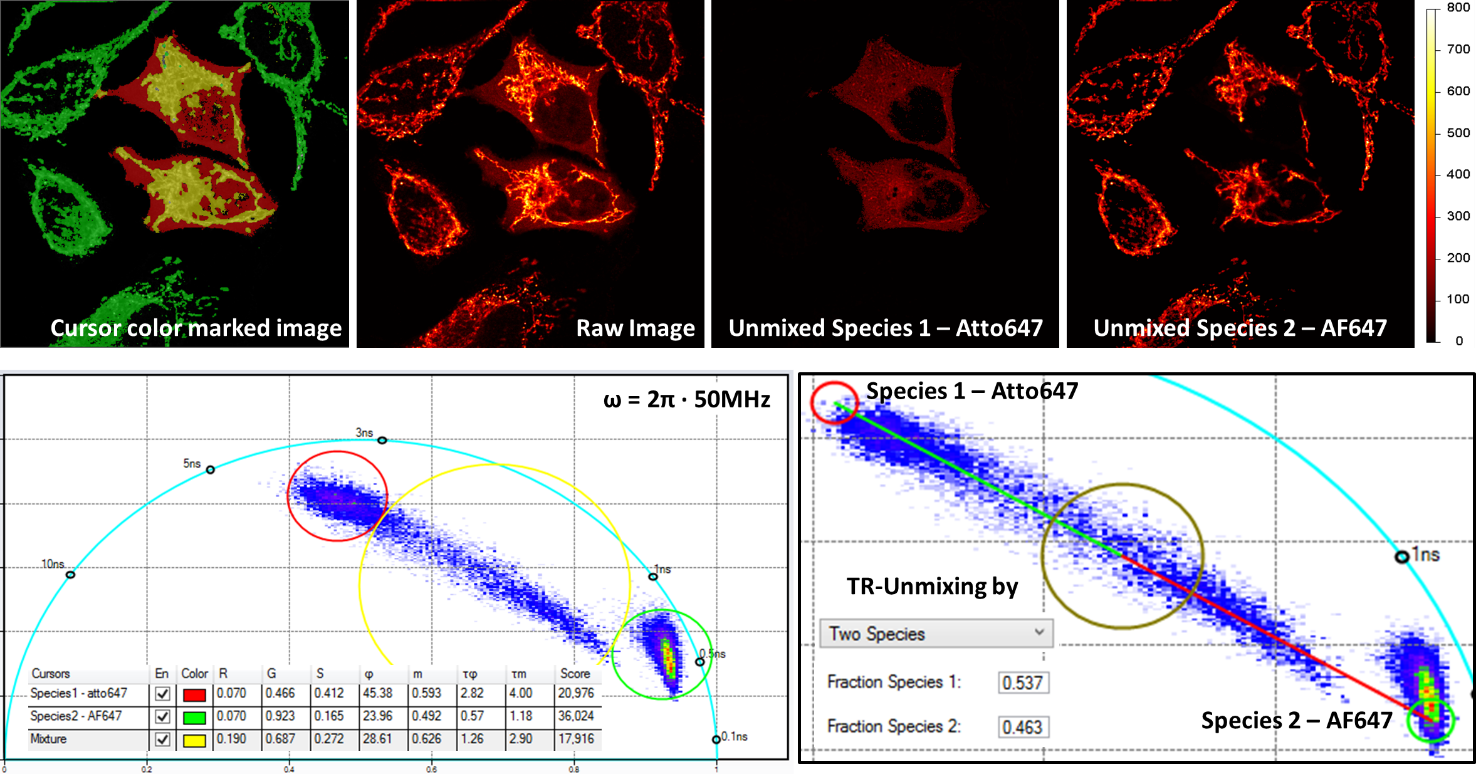
Multiplex imaging by time-resolved fluorescence requires using one excitation wavelength, and data is acquired on one detection channel. The figure shows the multiplexing of two labels, mitochondria labeled with Alexa Fluor 647 (AF647) and microtubule labeled with Atto 647 (Atto647) in the same cells. The raw image is acquired with excitation at 640 nm and detection in the same emission channel. Using the analysis routine in the phasor plot, the two structures are separated.
HomoFRET detection by polarization imaging
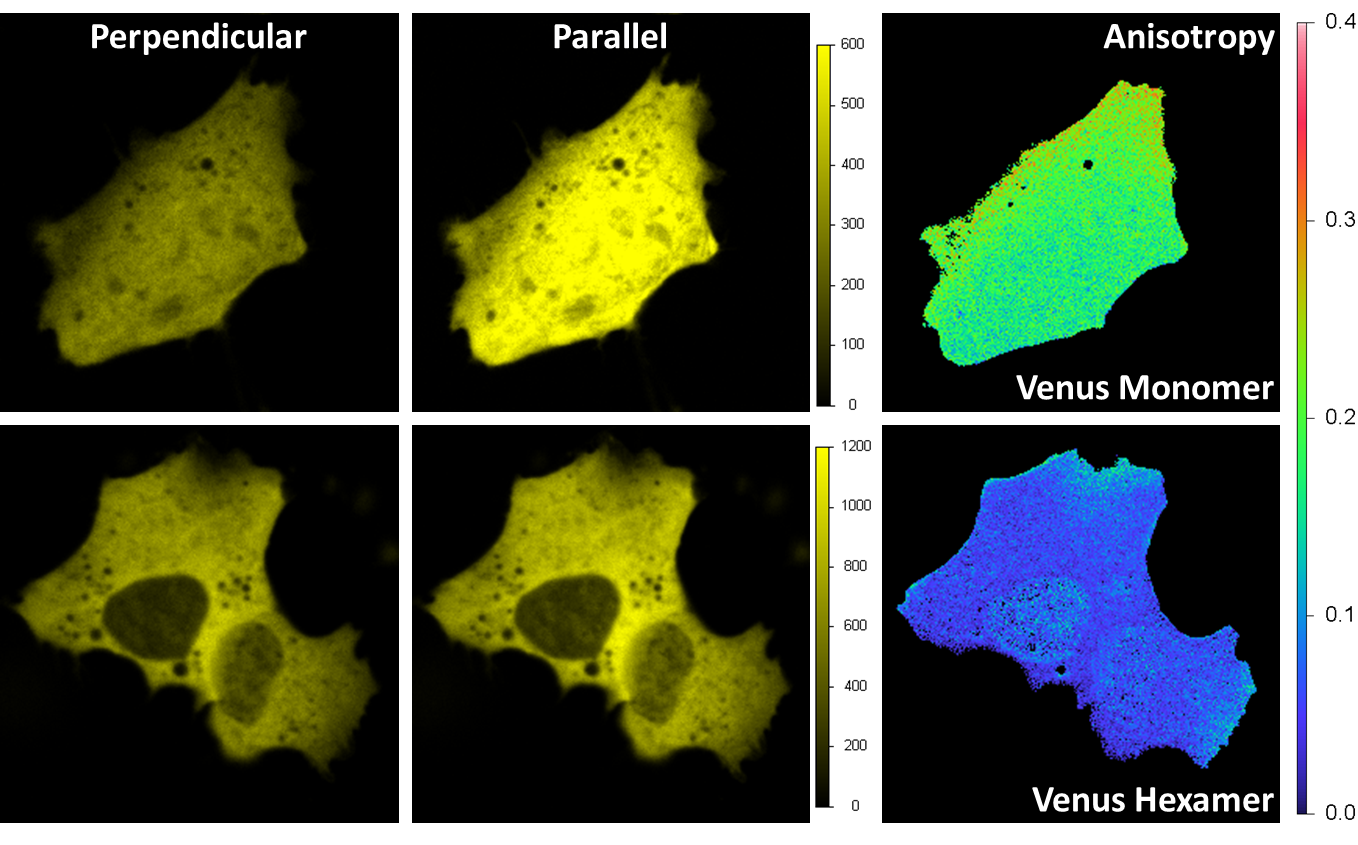
Venus was expressed in a HeLa cell. Excitation is 514 nm and emission is through a band-pass filter 545/35 nm. A beam-splitter polarizer in the emission splits the fluorescence beam into a perpendicular and parallel orientation beams, with each beam detected by one acquisition channel. The anisotropy image is calculated at each pixel. For the monomer (upper right) the average value of the anisotropy is around 0.2 indicating a prevalent orientation along the parallel axis; for the hexamer is less than 0.1 due to the local homo FRET between different Venus fluorescent proteins on the hexamer structure.
Single-molecule studies: Detection of single emitters by Antibunching
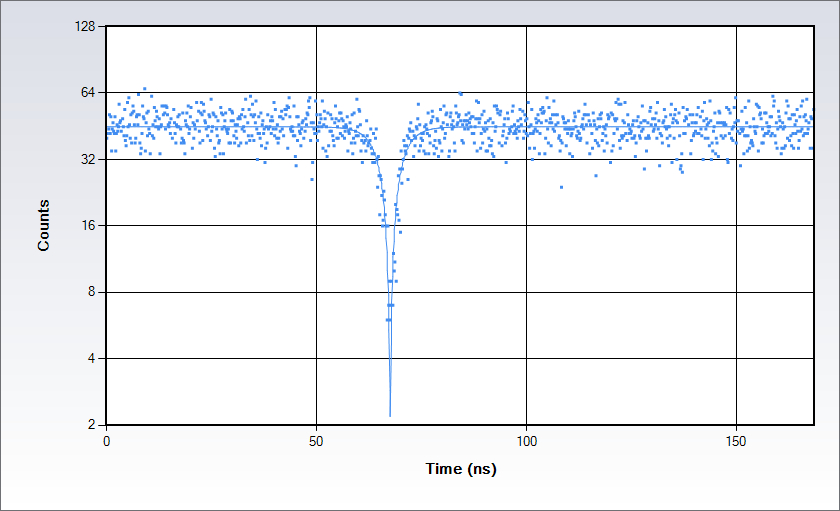
Antibunching data acquired on a solution of Rhodamine 110 in water with excitation at 488 nm. Measurements are acquired by splitting the signal in two beams following the classic Hanbury-Brown-Twiss set up and sending each of them to a separate detector of the Alba: the acquisition electronics provides a histogram of the time difference between the arrival time of the photons at the detectors. The histogram displays a dip at the coincidence point; the depth of the dip depends on the number of independent emitters in the observation volume while the shape depends on the excited state lifetime.
Product Specifications for Alba v5
Instrument Features
- Individual pinholes on each acquisition channel
- Computer-controlled selection of the pinhole variable aperture
- Computer-controlled positioning of the pinhole in the imaging plane
- Single-photon or multi-photon excitation
- Up to 4 channels data acquisition
- Auxiliary port for camera
Microscope
- Inverted and upright
Objectives
- Air objectives with 20X, 40X, 60X magnification and 1.5-8.1 working distances
- Oil immersion objectives, 1.4 NA and 60X (standard); other apertures available
- Water immersion objectives,1.2 NA 60X (standard), with coverslip correction (for 0.15-0.18 coverslip); other apertures available
Light Sources
- Single photon lasers housed in a laser launcher with computer-control of beam expander, laser intensity and shutter;
- Multi-photon excitation with computer-control of beam expander, laser intensity and shutter
Laser Launcher
- Models for 3-, 4-, 6-lasers. Light is delivered to the microscope through a single-mode fiber optic.
Galvanometer Scanner
- 2 silver-coated galvanometer scanning mirrors
- Clear optical surface: 3 mm
- Maximum scan rate: 5 KHz for 3 mm
- Scanning resolution: 64 x 64 to 4096 x 4096 pixels
- Scanning mode: Pt, Xt, XZ, XY, XZt, XYt, XYZ
- ROI scanning: rectangle, ellipse, polygon, line
Positioning Controls**
- ISS 3-axis control unit
- ISS XY galvo scanning mirrors control unit
- ISS Z-piezo control unit
- Microscope built-in focusing control module
- Automatic XY stages (ASI, Prior)
- XYZ piezo stages (MadCity, PI)
Pinhole
- Variable-aperture pinhole; diameter from 20 µm to 1000 µm
Detectors
- Cooled GaAsP and GaAs PMT
- Cooled Hybrid PMTs
- SPADs
Dichroic Filters
- For single-photon excitation: 1-, 2-, 3-band filters
- For multi-photon excitation
Polarizer
- Cube beam splitter, wavelength range: 450 - 1100 nm; extinction ratio: 10,000:1 at ±3 degrees
Data Acquisition Unit
- FastFLIM (Digital Frequency Domain FLIM)
- SWISS TCSPC card (Time Domain FLIM)
Software
- VistaVision
Computer & Monitor
- High-performance Processor, 32 GB RAM, Windows 11, 64-bit
- 32" monitor, 2556 x 1440 resolution
Power Requirements
- 110 - 240 V, 50/60 Hz, 400 VAC
Dimensions (mm)
- 885 (L) x 600 (W) x 330 (H)
Weight (kg)
- 40 (with no microscope)
Measurements for Alba v5
Intensity and Lifetime Imaging
- 1p or 2p confocal images
- FLIM in frequency-domain or in TCSPC
- Phosphorescence Lifetime Imaging (PLIM)
- Polarization images
Steady-state and Time-resolved Polarization Anisotropy Imaging
Fluorescence Fluctuations Spectroscopy (FFS)
- Fluorescence Correlation Spectroscopy (FCS)
- Fluorescence Cross-Correlation Spectroscopy (FCCS)
- Photon Counting Histogram (PCH)
- Fluorescence Lifetime Correlation Spectroscopy (FLCS)
- Scanning FCS
- Number & Brightness (N&B)
- Raster Imaging Correlation Spectroscopy (RICS)
Particle tracking (optional)
- 3D reconstruction of molecule trajectory
Super resolution (optional)
- Nanoimaging reconstruction with 20 nm resolution
Single Molecule FRET Bursts Analysis
- Burst Analysis
- FRET and Correlation methods
- PIE-FRET methods
Antibunching
Product Accessories for Alba v5
Additional Product Options
-
Perfusion system
A peristaltic pump supplies the stage with a solution for keeping the sample conditions (temperature, pH, etc...) stable.
-
Irrigation system
When using water objectives for prolonged measurements, it prevents the liquid drying up.
-
Auto Focus maintaining
It keeps the focus position of the objective for hours using an active feedback to counter drifts.
-
Sample temperature control
Stage top incubator or a full enclosure to maintain the environmental conditions of cell cultures.
-
Epifluorescence lamp
Visualize your sample with the Epi module. Select as light source either an arc lamp or an LED and the suitable filter cubes to add to the microscope cassette.
-
Atomic Force Microscope (AFM)
Fully integrated for the models:
NanoWizard by JPK-Bruker
Resolve by Bruker
For other models contact ISS.
Product Software for Alba v5
VistaVision
VistaVision is a complete software package for confocal microscopy applications including instrument control, data acquisition and data processing. Easy to use, the software has been developed in modular components that can be activated when a specific instrument configuration is selected. The modules include:
- FLIM/PLIM Imaging
- FFS
- smFRET
- Particle Tracking
Product Resources
-
Correlative confocal fluorescence lifetime and Atomic Force Microscopy imaging by ISS and JPK
-
FLIM Analysis using the Phasor Plots
-
Particle Tracking in a 2-Photon Excitation Microscope
-
The Sweet PIE (Pulsed Interleaved Excitation)
-
Using Alba with the FemtoFiber laser by Toptica for 2-photon quantitative imaging
-
FastFLIM STED for Alba v5
-
FLIM Analysis Enhanced with Phasor Plotting Aids Quantitative Biology
-
“Fluorescence lifetime imaging of physiological free Cu(ii) levels in live cells with a Cu(ii)-selective carbonic anhydrase-based biosensor.” Mccranor, B.J., Szmacinski, H., Zeng, H.H., Stoddard, A.K., Hurst, T., Fierke, C.A., Lakowicz, J.R. & Thompson, R.B. Metallomics, 6(5), p. 1034, 2014, Apr. doi: 10.1039/c3mt00305a.
-
“Application of Fluorescence Correlation Spectroscopy to Hapten–Antibody Binding.” Hazlett, T.L., Ruan, Q. & Tetin, S.Y. The Journal of Physical Chemistry B, 115(5), pp. 415–438, 2010, Dec. doi: 10.1385/1-59259-912-5:415.
-
“Determining Antibody Stoichiometry Using Time-Integrated Fluorescence Cumulant Analysis.” Skinner, J.P., Wu, B., Mueller, J.D. & Tetin, S.Y. The Journal of Physical Chemistry B, 115(5), pp. 1131–1138, 2010, Dec. doi: 10.1021/jp106279r.
-
“Fluorescence Correlation Spectroscopy Assay for Gliadin in Food.” Varriale, A., Rossi, M., Staiano, M., Terpetschnig, E., Barbieri, B., Rossi, M. & D'Auria, S. Analytical Chemistry, 79(12), pp. 4687–4689, 2007, May. doi: 10.1021/ac070475+.
-
“Antibodies in Diagnostic Applications.” Tetin, S. & Stroupe, S. Current Pharmaceutical Biotechnology, 5(1), pp. 9–16, 2004, Feb. doi: 10.2174/1389201043489602.
-
“Highly luminescent, biocompatible ytterbium(
iii ) complexes as near-infrared fluorophores for living cell imaging.” Ning, Y., Tang, J., Liu, Y.-W., Jing, J., Sun, Y. & Zhang, J.-L. Chemical Science, 9(15), pp. 3742–3753, 2018, May. doi: 10.1039/c8sc00259b. -
“Interleaflet Diffusion Coupling When Polymer Adsorbs onto One Sole Leaflet of a Supported Phospholipid Bilayer.” Zhang, L. & Granick, S. Macromolecules, 40(5), pp. 1366–1368, 2007, Feb. doi: 10.1021/ma062335s.
-
“How to Stabilize Phospholipid Liposomes (Using Nanoparticles).” Zhang, L. & Granick, S. Nano Letters, 6(4), pp. 694–698, 2006, Feb. doi: 10.1021/nl052455y.
-
“Slaved diffusion in phospholipid bilayers.” Zhang, L. & Granick, S. Proceedings of the National Academy of Sciences, 102(26), pp. 9118–9121, 2005, Jun. doi: 10.1073/pnas.0502723102.
-
“How Confined Lubricants Diffuse During Shear.” Mukhopadhyay, A., Bae, S.C., Zhao, J. & Granick, S. Physical Review Letters, 93(23), pp. 9118–9121, 2004, Dec. doi: 10.1103/physrevlett.93.236105.
-
“Polymer Lateral Diffusion at the Solid-Liquid Interface.” Zhao, J. & Granick, S. Journal of the American Chemical Society, 126(20), pp. 6242–6243, 2004, May. doi: 10.1021/ja0493749.
-
“Trapped Brownian Motion in Single- and Two-Photon Excitation Fluorescence Correlation Experiments.” Chirico, G., Fumagalli, C. & Baldini, G. The Journal of Physical Chemistry B, 106(10), pp. 2508–2519, 2002, Feb. doi: 10.1021/jp013087z.
-
“Stoichiometry of scaffold complexes in living neurons - DLC2 as a dimerization engine for GKAP.” Moutin, E., Compan, V., Raynaud, F., Clerté, C., Bouquier, N., Labesse, G., Ferguson, M.L., Fagni, L., Royer, C.A. & Perroy, J. Journal of Cell Science, 106(10), pp. 2508–2519, 2014, Jan. doi: 10.1242/jcs.145748.
-
“Reconciling molecular regulatory mechanisms with noise patterns of bacterial metabolic promoters in induced and repressed states.” Ferguson, M.L., Coq, D.L., Jules, M., Aymerich, S., Radulescu, O., Declerck, N. & Royer, C.A. Proceedings of the National Academy of Sciences, 109(1), pp. 155–160, 2011, Dec. doi: 10.1073/pnas.1110541108.
-
“Absolute quantification of gene expression in individual bacterial cells using two-photon fluctuation microscopy.” Ferguson, M.L., Coq, D.L., Jules, M., Aymerich, S., Declerck, N. & Royer, C.A. Analytical Biochemistry, 419(2), pp. 250–259, 2011, Dec. doi: 10.1016/j.ab.2011.08.017.
-
“Efficient Parallel Levenberg-Marquardt Model Fitting towards Real-Time Automated Parametric Imaging Microscopy.” Zhu, X. & Zhang, D. PLoS ONE, 8(10), p. e76665, 2013, Oct. doi: 10.1371/journal.pone.0076665.
-
“Nanometer-scale optical imaging of collagen fibers using gold nanoparticles.” Chen, B., Estrada, L.C., Hellriegel, C. & Gratton, E. Biomedical Optics Express, 2(3), p. 511, 2011, Feb. doi: 10.1364/boe.2.000511.
-
“Characterization of Brightness and Stoichiometry of Bright Particles by Flow-Fluorescence Fluctuation Spectroscopy.” Johnson, J., Chen, Y. & Mueller, J.D. Biophysical Journal, 99(9), pp. 3084–3092, 2010, Nov. doi: 10.1016/j.bpj.2010.08.057.
-
“Fluorescence correlation spectroscopy and photon counting histogram on membrane proteins: functional dynamics of the glycosylphosphatidylinositol-anchored urokinase plasminogen activator receptor.” Malengo, G., Andolfo, A., Sidenius, N., Gratton, E., Zamai, M. & Caiolfa, V.R. Journal of Biomedical Optics, 13(3), p. 031215, 2008, Nov. doi: 10.1117/1.2940570.
-
“Unraveling Protein-Protein Interactions in Living Cells with Fluorescence Fluctuation Brightness Analysis.” Chen, Y., Wei, L.-N. & Müller, J.D. Biophysical Journal, 88(6), pp. 4366–4377, 2005, Jun. doi: 10.1529/biophysj.105.059170.
-
“Dual-Color Photon-Counting Histogram.” Chen, Y., Tekmen, M., Hillesheim, L., Skinner, J., Wu, B. & Müller, J.D. Biophysical Journal, 88(3), pp. 2177–2192, 2005, Mar. doi: 10.1529/biophysj.104.048413.
-
“Fluorescence Spectroscopy with Metal–Dielectric Waveguides.” Badugu, R., Szmacinski, H., Ray, K., Descrovi, E., Ricciardi, S., Zhang, D., Chen, J., Huo, Y. & Lakowicz, J.R. The Journal of Physical Chemistry C, 119(28), pp. 16245–16255, 2015, Jul. doi: 10.1021/acs.jpcc.5b04204.
-
“Imaging of Protein Secretion from a Single Cell Using Plasmonic Substrates.” Szmacinski, H., Toshchakov, V., Piao, W. & Lakowicz, J.R. BioNanoScience, 3(1), pp. 30–36, 2013, Jan. doi: 10.1007/s12668-013-0076-7.
-
“Confocal Fluctuation Spectroscopy and Imaging.” Foldes-Papp, Z., Liao, S.-C.J., You, T., Terpetschnig, E. & Barbieri, B. Current Pharmaceutical Biotechnology, 11(6), pp. 639–653, 2010, Sep. doi: 10.2174/138920110792246618.
-
“Fluorescence fluctuation spectroscopy: ushering in a new age of enlightenment for cellular dynamics.” Jameson, D.M., Ross, J.A. & Albanesi, J.P. Biophysical Reviews, 1(3), pp. 105–118, 2009, Aug. doi: 10.1007/s12551-009-0013-8.
-
“Capturing directed molecular motion in the nuclear pore complex of live cells.” Cardarelli, F., Lanzano, L. & Gratton, E. Proceedings of the National Academy of Sciences, 109(25), pp. 9863–9868, 2012, Jun. doi: 10.1073/pnas.1200486109.
-
“Nanometer-scale imaging by the modulation tracking method.” Lanzano, L., Digman, M.A., Fwu, P., Giral, H., Levi, M. & Gratton, E. Journal of Biophotonics, 4(6), pp. 415–424, 2011, Apr. doi: 10.1002/jbio.201100002.
-
“A Zn-dependent structural transition of SOD1 modulates its ability to undergo phase separation.” Das, B., Roychowdhury, S., Mohanty, P., Rizuan, A., Chakraborty, J., Mittal, J. & Chattopadhyay, K. The EMBO Journal, 42(2), 2022, Nov. doi: 10.15252/embj.2022111185.
-
“Chromosome loading of cohesin depends on conserved residues in Scc3.” Pathania, A., Liu, W., Matityahu, A., Irudayaraj, J. & Onn, I. Current Genetics, 67(3), pp. 447–459, 2021, Jan. doi: 10.1007/s00294-020-01150-3.
-
“CAF-1 and Rtt101p function within the replication-coupled chromatin assembly network to promote H4 K16ac, preventing ectopic silencing.” Young, T.J., Cui, Y., Pfeffer, C., Hobbs, E., Liu, W., Irudayaraj, J. & Kirchmaier, A.L. PLOS Genetics, 16(12), p. e1009226, 2020, Dec. doi: 10.1371/journal.pgen.1009226.
-
“Copper mediates mitochondrial biogenesis in retinal pigment epithelial cells.” Dhivya, M.A., Aberami, S., Nikhalashree, S., Biswas, J., Liu, W., Irudayaraj, J., Sulochana, K., Coral, K. & Devi, S.B. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease, 1866(10), p. 165843, 2020, Oct. doi: 10.1016/j.bbadis.2020.165843.
-
“Quantitative High-Resolution Imaging of Live Microbial Cells at High Hydrostatic Pressure.” Bourges, A.C., Lazarev, A., Declerck, N., Rogers, K.L. & Royer, C.A. Biophysical Journal, 118(11), pp. 2670–2679, 2020, Jun. doi: 10.1016/j.bpj.2020.04.017.
-
“Monomeric cohesin state revealed by live-cell single-molecule spectroscopy.” Liu, W., Biton, E., Pathania, A., Matityahu, A., Irudayaraj, J. & Onn, I. EMBO reports, 21(2), 2019, Dec. doi: 10.15252/embr.201948211.
-
“Epigenetic biomarker screening by FLIM-FRET for combination therapy in ER+ breast cancer.” Liu, W., Cui, Y., Ren, W. & Irudayaraj, J. Clinical Epigenetics, 11(1), 2019, Jan. doi: 10.1186/s13148-019-0620-6.
-
“The N-Terminal Domain of ALS-Linked TDP-43 Assembles without Misfolding.” Tsoi, P.S., Choi, K.-J., Leonard, P.G., Sizovs, A., Moosa, M.M., Mackenzie, K.R., Ferreon, J.C. & Ferreon, A.C.M. Angewandte Chemie International Edition, 56(41), pp. 12590–12593, 2017, Sep. doi: 10.1002/anie.201706769.
-
“The Use of FLIM-FRET for the Detection of Mitochondria-Associated Protein Interactions.” Osterlund, E.J., Liu, Q. & Andrews, D.W. Angewandte Chemie International Edition, 56(41), pp. 395–419, 2015, Sep. doi: 10.1007/978-1-4939-2257-4_34.
-
“A Decoy Peptide that Disrupts TIRAP Recruitment to TLRs Is Protective in a Murine Model of Influenza.” Piao, W., Shirey, K., Ru, L., Lai, W., Szmacinski, H., Snyder, G., Sundberg, E., Lakowicz, J., Vogel, S. & Toshchakov, V. Cell Reports, 11(12), pp. 1941–1952, 2015, Jun. doi: 10.1016/j.celrep.2015.05.035.
-
“Measuring Förster Resonance Energy Transfer Using Fluorescence Lifetime Imaging Microscopy.” Day, R.N. Microscopy Today, 23(3), pp. 44–51, 2015, Apr. doi: 10.1017/s1551929515000395.
-
“Binding of fusion protein FLSC IgG1 to CCR5 is enhanced by CCR5 antagonist Maraviroc.” Latinovic, O., Schneider, K., Szmacinski, H., Lakowicz, J.R., Heredia, A. & Redfield, R.R. Antiviral Research, 112(5), pp. 80–90, 2014, Dec. doi: 10.1016/j.antiviral.2014.10.006.
-
“Cis and trans internucleosomal interactions of H3 and H4 tails in tetranucleosomes.” Nurse, N.P. & Yuan, C. Biopolymers, 103(1), pp. 33–40, 2014, Oct. doi: 10.1002/bip.22560.
-
“DNA Methylation Effects on Tetra-Nucleosome Compaction and Aggregation.” Jimenez-Useche, I., Nurse, N., Tian, Y., Kansara, B., Shim, D. & Yuan, C. Biophysical Journal, 107(7), pp. 1629–1636, 2014, Oct. doi: 10.1016/j.bpj.2014.05.055.
-
“Application of phasor plot and autofluorescence correction for study of heterogeneous cell population.” Szmacinski, H., Toshchakov, V. & Lakowicz, J.R. Journal of Biomedical Optics, 19(4), p. 046017, 2014, Apr. doi: 10.1117/1.jbo.19.4.046017.
-
“mMAPS: A Flow-Proteometric Technique to Analyze Protein-Protein Interactions in Individual Signaling Complexes.” Chou, C.-K., Lee, H.-H., Tsou, P.-H., Chen, C.-T., Hsu, J.-M., Yamaguchi, H., Wang, Y.-N., Lee, H.-J., Hsu, J.L., Lee, J.-F., Kameoka, J. & Hung, M.-C. Science Signaling, 7(315), pp. 517–524, 2014, Mar. doi: 10.1126/scisignal.2004620.
-
“Unexpected Complex Formation between Coralyne and Cyclic Diadenosine Monophosphate Providing a Simple Fluorescent Turn-on Assay to Detect This Bacterial Second Messenger.” Zhou, J., Sayre, D.A., Zheng, Y., Szmacinski, H. & Sintim, H.O. Analytical Chemistry, 86(5), pp. 2412–2420, 2014, Feb. doi: 10.1021/ac403203x.
-
“Unmethylated and methylated CpG dinucleotides distinctively regulate the physical properties of DNA.” Jimenez-Useche, I., Shim, D., Yu, J. & Yuan, C. Biopolymers, 101(5), pp. 517–524, 2014, Feb. doi: 10.1002/bip.22411.
-
“Unraveling transcription factor interactions with heterochromatin protein 1 using fluorescence lifetime imaging microscopy and fluorescence correlation spectroscopy.” Siegel, A.P., Hays, N.M. & Day, R.N. Journal of Biomedical Optics, 18(2), p. 025002, 2013, Feb. doi: 10.1117/1.jbo.18.2.025002.
-
“Monitoring Biosensor Activity in Living Cells with Fluorescence Lifetime Imaging Microscopy.” Hum, J., Siegel, A., Pavalko, F. & Day, R. International Journal of Molecular Sciences, 13(12), pp. 14385–14400, 2012, Nov. doi: 10.3390/ijms131114385.
-
“Chromophore maturation and fluorescence fluctuation spectroscopy of fluorescent proteins in a cell-free expression system.” Macdonald, P.J., Chen, Y. & Mueller, J.D. Analytical Biochemistry, 421(1), pp. 291–298, 2012, Feb. doi: 10.1016/j.ab.2011.10.040.
-
“HIV-1 Nef Binds a Subpopulation of MHC-I throughout Its Trafficking Itinerary and Down-regulates MHC-I by Perturbing Both Anterograde and Retrograde Trafficking.” Yi, L., Rosales, T., Rose, J.J., Chaudhury, B., Knutson, J.R. & Venkatesan, S. Journal of Biological Chemistry, 285(40), pp. 30884–30905, 2010, Oct. doi: 10.1074/jbc.m110.135947.
-
“Familial Hypertrophic Cardiomyopathy Can Be Characterized by a Specific Pattern of Orientation Fluctuations of Actin Molecules,.” Borejdo, J., Szczesna-Cordary, D., Muthu, P. & Calander, N. Biochemistry, 49(25), pp. 5269–5277, 2010, Jun. doi: 10.1021/bi1006749.
-
“Estrogen Receptor Interactions and Dynamics Monitored in Live Cells by Fluorescence Cross-Correlation Spectroscopy.” Savatier, J., Jalaguier, S., Ferguson, M.L., Cavaillès, V. & Royer, C.A. Biochemistry, 49(4), pp. 772–781, 2010, Jan. doi: 10.1021/bi9013006.
-
“Monomer-Dimer Equilibrium in Glutathione Transferases: A Critical Re-Examination.” Fabrini, R., Luca, A.D., Stella, L., Mei, G., Orioni, B., Ciccone, S., Federici, G., Bello, M.L. & Ricci, G. Biochemistry, 48(43), pp. 10473–10482, 2009, Oct. doi: 10.1021/bi901238t.
-
“A Fluorescent Mutant of the NM Domain of the Yeast Prion Sup35 Provides Insight into Fibril Formation and Stability.” Palhano, F.L., Rocha, C.B., Bernardino, A., Weissmuller, G., Masuda, C.A., Montero-Lomelí, M., Gomes, A.M., Chien, P., Fernandes, P.M.B. & Foguel, D. Biochemistry, 48(29), pp. 6811–6823, 2009, Jul. doi: 10.1021/bi9000276.
-
“Fluorescence Correlation Spectroscopy of Phosphatidylinositol-Specific Phospholipase C Monitors the Interplay of Substrate and Activator Lipid Binding.” Pu, M., Roberts, M.F. & Gershenson, A. Biochemistry, 48(29), pp. 6835–6845, 2009, Jul. doi: 10.1021/bi900633p.
-
“Correlation of Vesicle Binding and Phospholipid Dynamics with Phospholipase C Activity.” Pu, M., Fang, X., Redfield, A.G., Gershenson, A. & Roberts, M.F. Journal of Biological Chemistry, 284(24), pp. 16099–16107, 2009, Jun. doi: 10.1074/jbc.m809600200.
-
“Characterization of the Control Catabolite Protein of Gluconeogenic Genes Repressor by Fluorescence Cross-Correlation Spectroscopy and Other Biophysical Approaches.” Zorrilla, S., Ortega, Á., Chaix, D., Alfonso, C., Rivas, G., Aymerich, S., Lillo, M.P., Declerck, N. & Royer, C.A. Biophysical Journal, 95(9), pp. 4403–4415, 2008, Nov. doi: 10.1529/biophysj.108.135863.
-
“Endothelial adhesion receptors are recruited to adherent leukocytes by inclusion in preformed tetraspanin nanoplatforms.” Barreiro, O., Zamai, M., Yáñez-Mó, M., Tejera, E., López-Romero, P., Monk, P.N., Gratton, E., Caiolfa, V.R. & Sánchez-Madrid, F. Journal of Cell Biology, 183(3), pp. 527–542, 2008, Oct. doi: 10.1083/jcb.200805076.
-
“Applications of dual-color fluorescence cross-correlation spectroscopy in antibody binding studies.” Ruan, Q. & Tetin, S.Y. Analytical Biochemistry, 374(1), pp. 182–195, 2008, Mar. doi: 10.1016/j.ab.2007.11.007.
-
“Fructose-1,6-bisphosphate Acts Both as an Inducer and as a Structural Cofactor of the Central Glycolytic Genes Repressor (CggR).” Zorrilla, S., Chaix, D., Ortega, A., Alfonso, C., Doan, T., Margeat, E., Rivas, G., Aymerich, S., Declerck, N. & Royer, C.A. Biochemistry, 46(51), pp. 14996–15008, 2007, Dec. doi: 10.1021/bi701805e.
-
“Monomer–dimer dynamics and distribution of GPI-anchored uPAR are determined by cell surface protein assemblies.” Caiolfa, V.R., Zamai, M., Malengo, G., Andolfo, A., Madsen, C.D., Sutin, J., Digman, M.A., Gratton, E., Blasi, F. & Sidenius, N. Journal of Cell Biology, 179(5), pp. 1067–1082, 2007, Dec. doi: 10.1083/jcb.200702151.
-
“Escherichia coli ribosomal protein L20 binds as a single monomer to its own mRNA bearing two potential binding sites.” Allemand, F., Haentjens, J., Chiaruttini, C., Royer, C. & Springer, M. Nucleic Acids Research, 35(9), pp. 3016–3031, 2007, Apr. doi: 10.1093/nar/gkm197.
-
“Determining the stoichiometry of protein heterocomplexes in living cells with fluorescence fluctuation spectroscopy.” Chen, Y. & Müller, J.D. Proceedings of the National Academy of Sciences, 104(9), pp. 3147–3152, 2007, Feb. doi: 10.1073/pnas.0606557104.
-
“Interactions of Two Monoclonal Antibodies with BNP: High Resolution Epitope Mapping Using Fluorescence Correlation Spectroscopy.” Tetin, S.Y., Ruan, Q., Saldana, S.C., Pope, M.R., Chen, Y., Wu, H., Pinkus, M.S., Jiang, J. & Richardson, P.L. Biochemistry, 45(47), pp. 14155–14165, 2006, Nov. doi: 10.1021/bi0607047.
-
“Quantitative detection of the ligand-dependent interaction between the androgen receptor and the co-activator, Tif2, in live cells using two color, two photon fluorescence cross-correlation spectroscopy.” Rosales, T., Georget, V., Malide, D., Smirnov, A., Xu, J., Combs, C., Knutson, J.R., Nicolas, J.-C. & Royer, C.A. European Biophysics Journal, 36(2), pp. 153–161, 2006, Oct. doi: 10.1007/s00249-006-0095-1.
-
“Effects of Protein-Ligand Associations on the Subunit Interactions of Phosphofructokinase from B. stearothermophilus.” Quinlan, R.J. & Reinhart, G.D. Biochemistry, 45(38), pp. 11333–11341, 2006, Sep. doi: 10.1021/bi0608921.
-
“Phosphoinositide Specificity of and Mechanism of Lipid Domain Formation by Annexin A2-p11 Heterotetramer.” Gokhale, N.A., Abraham, A., Digman, M.A., Gratton, E. & Cho, W. Journal of Biological Chemistry, 280(52), pp. 42831–42840, 2005, Dec. doi: 10.1074/jbc.m508129200.
-
“The Nuclear Receptor Coactivator PGC-1α Exhibits Modes of Interaction with the Estrogen Receptor Distinct From those of SRC-1.” Bourdoncle, A., Labesse, G., Margueron, R., Castet, A., Cavaillès, V. & Royer, C.A. Journal of Molecular Biology, 347(5), pp. 921–934, 2005, Apr. doi: 10.1016/j.jmb.2005.01.048.
-
“Polarized fluorescence correlation spectroscopy of DNA-DAPI complexes.” Barcellona, M.L., Gammon, S., Hazlett, T., Digman, M.A. & Gratton, E. Microscopy Research and Technique, 65(4-5), pp. 205–217, 2004, Nov. doi: 10.1002/jemt.20121.
-
“Vesicle Encapsulation Studies Reveal that Single Molecule Ribozyme Heterogeneities Are Intrinsic.” Okumus, B., Wilson, T.J., Lilley, D.M. & Ha, T. Biophysical Journal, 87(4), pp. 2798–2806, 2004, Oct. doi: 10.1529/biophysj.104.045971.
-
“Pleckstrin Homology Domain Diffusion in Dictyostelium Cytoplasm Studied Using Fluorescence Correlation Spectroscopy.” Ruchira, , Hink, M.A., Bosgraaf, L., Haastert, P.J.v. & Visser, A.J. Journal of Biological Chemistry, 279(11), pp. 10013–10019, 2004, Mar. doi: 10.1074/jbc.m310039200.
-
“Probing protein oligomerization in living cells with fluorescence fluctuation spectroscopy.” Chen, Y., Wei, L.-N. & Müller, J.D. Proceedings of the National Academy of Sciences, 100(26), pp. 15492–15497, 2003, Dec. doi: 10.1073/pnas.2533045100.
-
“Segregation of Saturated Chain Lipids in Pulmonary Surfactant Filmsand Bilayers.” Nag, K., Pao, J.-S., Harbottle, R.R., Possmayer, F., Petersen, N.O. & Bagatolli, L.A. Biophysical Journal, 82(4), pp. 2041–2051, 2002, Apr. doi: 10.1016/s0006-3495(02)75552-5.
-
“Molecular Heterogeneity of O-Acetylserine Sulfhydrylase by Two-Photon Excited Fluorescence Fluctuation Spectroscopy.” Chirico, G., Bettati, S., Mozzarelli, A., Chen, Y., Müller, J.D. & Gratton, E. Biophysical Journal, 80(4), pp. 1973–1985, 2001, Apr. doi: 10.1016/s0006-3495(01)76167-x.
-
“Single-Molecule Tracking in Live Cell without Immobilization or without Hydrodynamic Flow by Simulations: Thermodynamic Jitter.” Baumann, G., and Földes-Papp, Z. Biophysica, 4, p. 442-452, 2024, Aug. doi: 10.3390/e22121380.
-
“Study on Single-molecule Biophysics and Biochemistry in Dilute Liquids and Live Cells without Immobilization or Significant Hydrodynamic Flow: The Thermodynamic Single-molecule Demon.” Foldes-Papp, Z. & Baumann, G. Current Pharmaceutical Biotechnology, 23(14), pp. 1750–1757, 2022, Nov. doi: 10.2174/1389201023666220616123928.
-
“Single-molecule time resolution in dilute liquids and live cells at the molecular scale: Constraints on the measurement time.” Földes-Papp, Z. American Journal of Translational Medicine, 5(3), pp. 154 - 165, 2021, Jan.
-
“Visualization of subdiffusive sites in a live single cell.” Földes-Papp, Z., Baumann, G. & Li, L.-C. Journal of Biological Methods, 8(1), p. e142, 2021, Jan. doi: 10.14440/jbm.2021.348.
-
“Measurements of Single Molecules in Solution and Live Cells Over Longer Observation Times Than Those Currently Possible: The Meaningful Time.” Foldes-Papp, Z. Current Pharmaceutical Biotechnology, 14(4), pp. 441–444, 2013, Apr. doi: 10.2174/1389201011314040009.
-
“Fluorescence Molecule Counting for Single-Molecule Studies in Crowded Environment of Living Cells without and with Broken Ergodicity.” Foldes-Papp, Z. & Baumann, G. Current Pharmaceutical Biotechnology, 12(5), pp. 824–833, 2011, May. doi: 10.2174/138920111795470949.
-
“Single actomyosin motor interactions in skeletal muscle.” Földes-Papp, Z., Liao, S.-C.J., Barbieri, B., Gryczynski, K., Luchowski, R., Gryczynski, Z., Gryczynski, I., Borejdo, J. & You, T. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, 1813(5), pp. 858–866, 2011, May. doi: 10.1016/j.bbamcr.2011.02.001.
-
“Meaningful Interpretation of Subdiffusive Measurements in Living Cells (Crowded Environment) by Fluorescence Fluctuation Microscopy.” Baumann, G., Place, R.F. & Foldes-Papp, Z. Current Pharmaceutical Biotechnology, 11(5), pp. 527–543, 2010, Aug. doi: 10.2174/138920110791591454.
-
“Anomalous behavior in length distributions of 3D random Brownian walks and measured photon count rates within observation volumes of single-molecule trajectories in fluorescence fluctuation microscopy.” Baumann, G., Gryczynski, I. & Földes-Papp, Z. Optics Express, 18(17), p. 17883, 2010, Aug. doi: 10.1364/oe.18.017883.
-
“Editorial [Hot Topic: Getting High on Single Molecule Biophysics (Guest Editor: Steven M. Block )].” Block, S. Current Pharmaceutical Biotechnology, 10(5), pp. 464–466, 2009, Aug. doi: 10.2174/138920109788922092.
-
“Reducing Background Contributions in Fluorescence Fluctuation Time- Traces for Single-Molecule Measurements in Solution.” Foldes-Papp, Z., Liao, S.-C., You, T. & Barbieri, B. Current Pharmaceutical Biotechnology, 10(5), pp. 532–542, 2009, Aug. doi: 10.2174/138920109788922137.
-
“Fluorescence Fluctuation Spectroscopic Approaches to the Study of a Single Molecule Diffusing in Solution and a Live Cell without Systemic Drift or Convection: A Theoretical Study.” Foldes-Papp, Z. Current Pharmaceutical Biotechnology, 8(5), pp. 261–273, 2007, Oct. doi: 10.2174/138920107782109930.
-
“Mobility and distribution of replication protein A in living cells using fluorescence correlation spectroscopy.” Braet, C., Stephan, H., Dobbie, I.M., Togashi, D.M., Ryder, A.G., Földes-Papp, Z., Lowndes, N. & Nasheuer, H.P. Experimental and Molecular Pathology, 82(2), pp. 156–162, 2007, Apr. doi: 10.1016/j.yexmp.2006.12.008.
-
“`True' single-molecule molecule observations by fluorescence correlation spectroscopy and two-color fluorescence cross-correlation spectroscopy.” Földes-Papp, Z. Experimental and Molecular Pathology, 82(2), pp. 147–155, 2007, Apr. doi: 10.1016/j.yexmp.2006.12.002.
-
“What it means to measure a single molecule in a solution by fluorescence fluctuation spectroscopy.” Földes-Papp, Z. Experimental and Molecular Pathology, 80(3), pp. 209–218, 2006, Jun. doi: 10.1016/j.yexmp.2006.01.001.
-
“How the Molecule Number Is Correctly Quantified in Two-Color Fluorescence Cross-Correlation Spectroscopy: Corrections for Cross-Talk and Quenching in Experiments.” Foldes-Papp, Z. Current Pharmaceutical Biotechnology, 6(6), pp. 437–444, 2005, Dec. doi: 10.2174/138920105775159296.
-
“Single-Phase Single-Molecule Fluorescence Correlation Spectroscopy (SPSM-FCS).” Földes-Papp, Z., Fuchs, J. & Podda, M. Encyclopedia of Medical Genomics and Proteomics, 1, pp. 1–7, 2004, Dec.
-
“Detection of single molecules: solution-phase single-molecule fluorescence correlation spectroscopy as an ultrasensitive, rapid and reliable system for immunological investigation.” Földes-Papp, Z., Demel, U. & Tilz, G.P. Journal of Immunological Methods, 260(1-2), pp. 117–124, 2002, Feb. doi: 10.1016/s0022-1759(01)00537-3.
Configurations of Alba v5
Alba can be selected in either one of two geometric configurations: the branch configuration where the fluorescence beams for each channel are separated in cascade, and the parallel configuration where the fluorescence beams are separate for the channels 1-2 and 3-4, respectively.

























