
Overview of Alba v5
Alba v5是一个结合了各种测量模式的激光扫描显微镜,在实验量化生物学和材料科学中对单分子探测灵敏度有较高要求的情况下应用。它可以从光谱中的紫色区域至近红外区域进行采集,带有两个独立的激光入射端口。Alba v5由VistaVision驱动。VistaVision是一款综合性软件包,用于仪器控制、图像获取及处理。
Alba v5的关键特征
多模态成像 (强度、寿命和SHG)
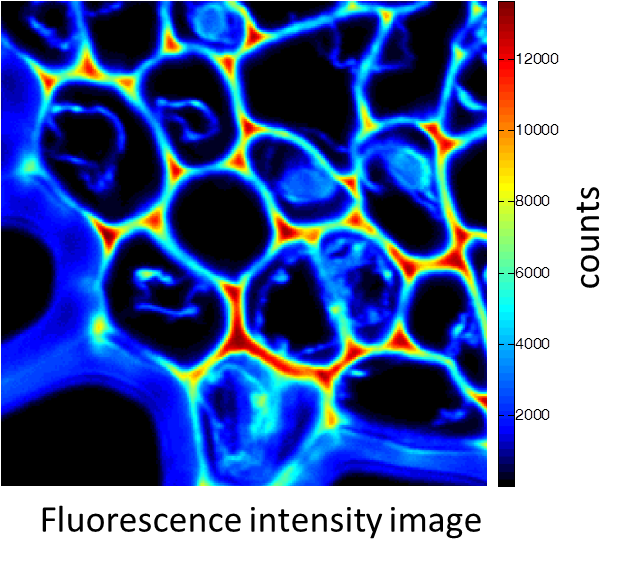
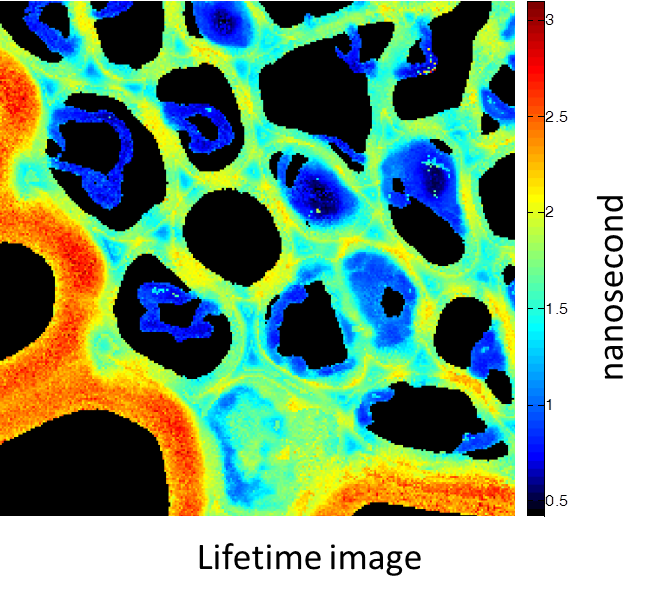
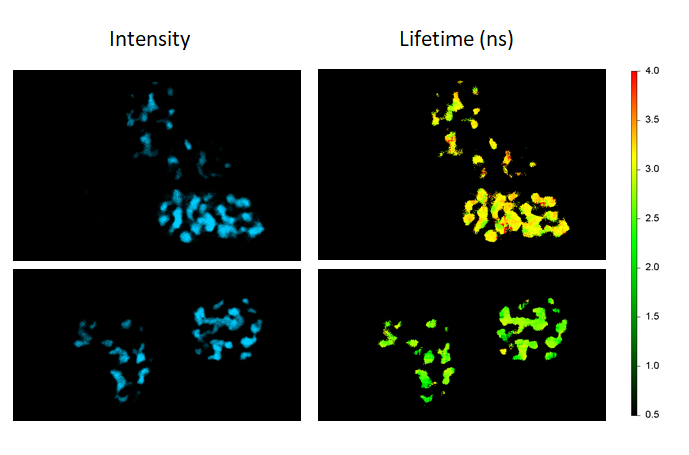
铃兰的根茎横截面。图像由带有多光子激光器 (在800 nm处发射) 的Alba获得。标准荧光强度图像 (图1) 荧光寿命图像 (图2) 和二次谐波生成 (SHG) 图像 (图3)。 所有的图像都是256 × 256像素、40 × 40 µm2。
(由伊利诺伊厄巴纳贝克曼研究所的zhang博士提供)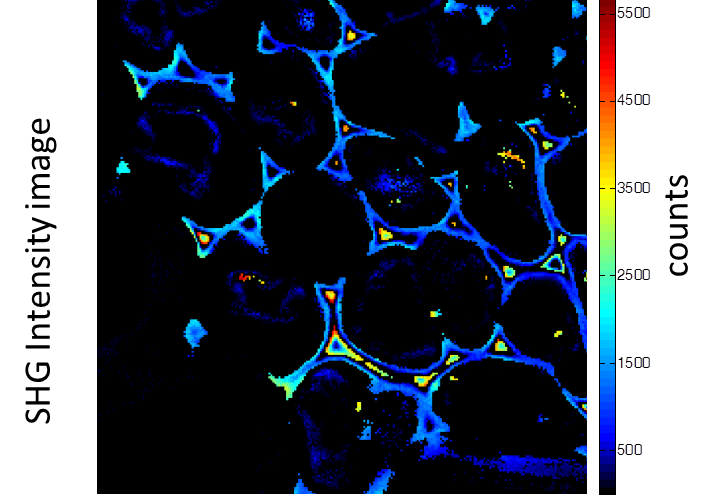
组织成像
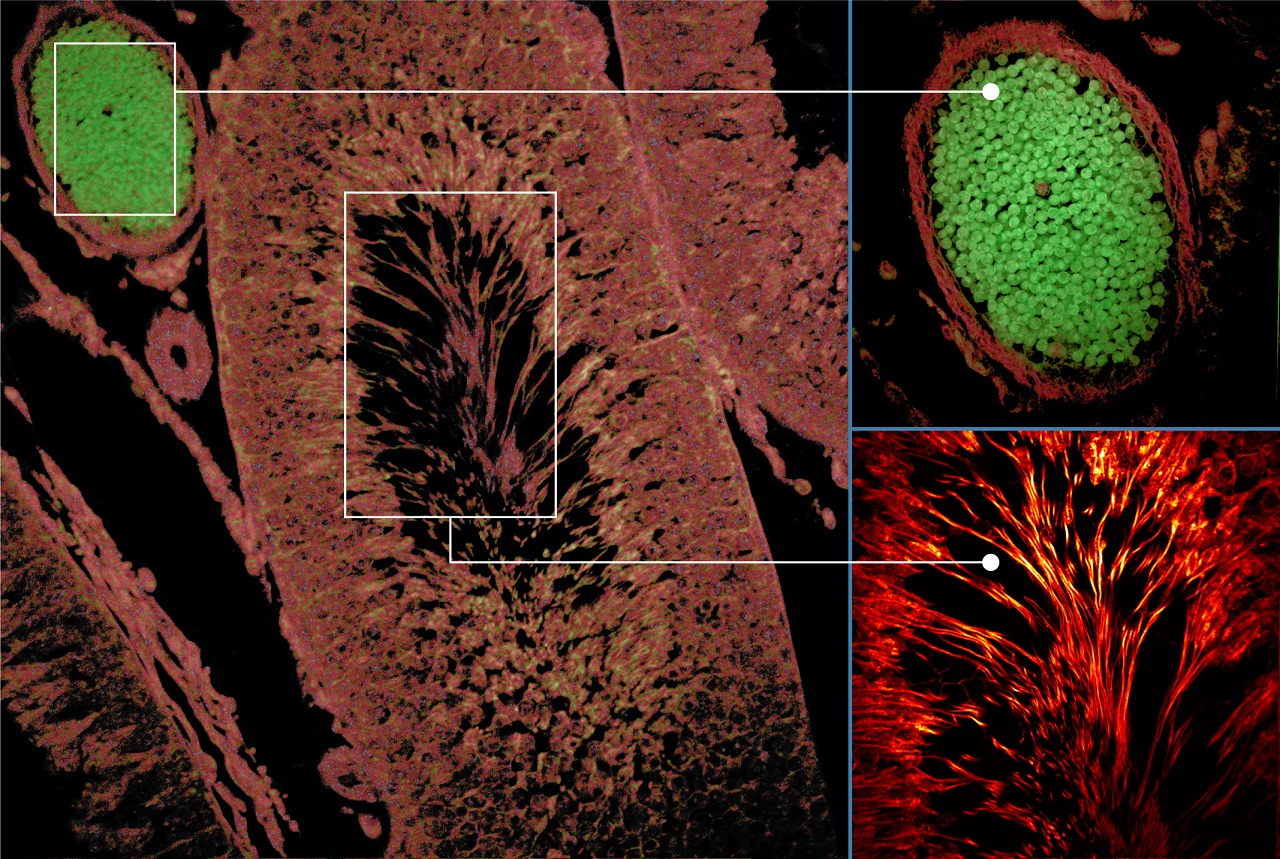
自发荧光组织图像:激发:375 nm,发射:425 nm,物镜:20X NA0.75
(由GSK的Aneesh Alex博士提供)
活细胞中的FLIM-FRET
FLIM-FRET绘制了活细胞中共同表达Cerulean和Venus的Cebp/α 蛋白质FRET效率,以此在细胞核 (仅供体控制:仅表达Cebp/α-Cerulean的细胞;FRET样本:共同表达Cebp/α-Cerulean和Cebp/α-Venus的细胞)定位其二聚化
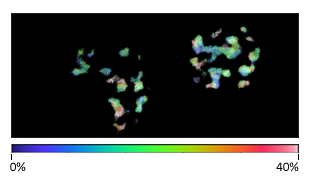
使用相位图通过时间分辨解混合获得的多重成像
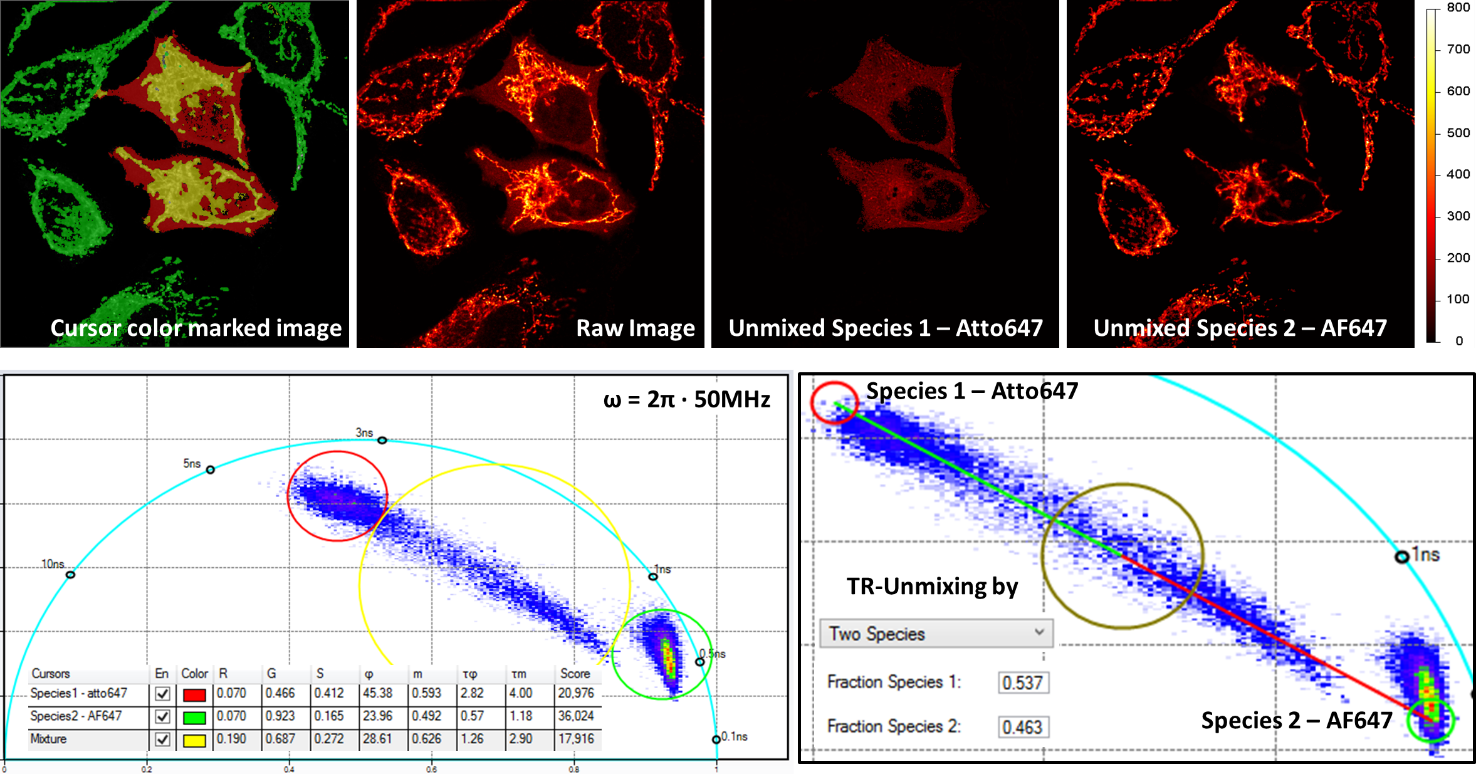
通过时间分辨荧光进行的多重成像需要使用单个激发波长,并且数据是在单个探测通道上获取的。上图显示了双标记的多重成像,同一细胞内用Alexa Fluor 647 (AF647) 标记的线粒体和Atto 647 (Atto647) 标记的微管。原始图像获取时是在640 nm处激发的,并且探测是在同一发射通道中进行的。利用相位图中的分析程式,两个结构可以被分开。
通过偏振成像实现的HomoFRET探测
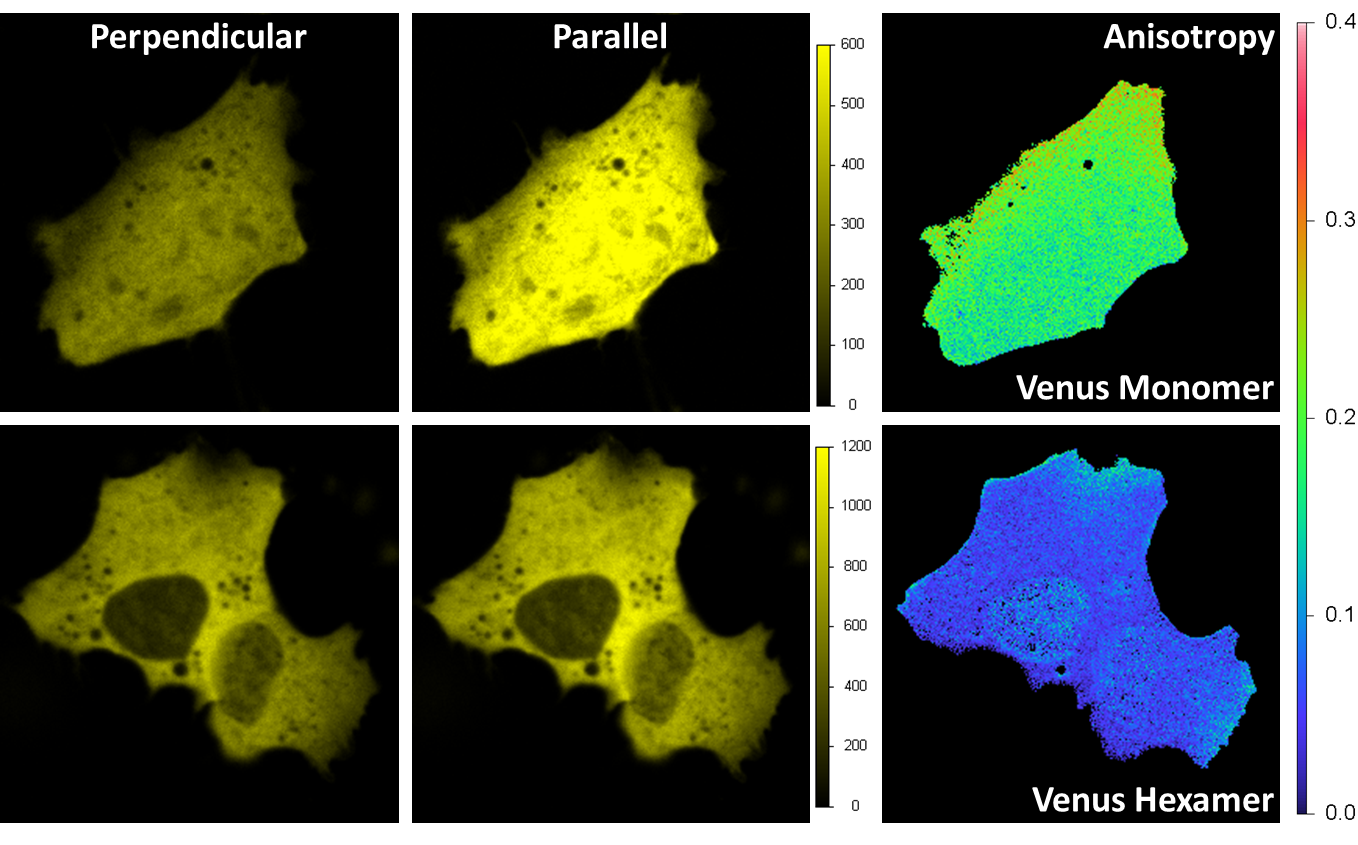
Venus在海拉细胞内被表达。激发波长为514 nm,并且发射是通过带通过滤器545/35 nm完成的。发射中的分束偏振器会将荧光光束分成垂直和平行方向的光束,每一个光束由一个采集通道进行探测。各向异性图像在各个像素上被算出。对于单体 (右上) 来说,各向异性的平均值约为0.2,这说明了普遍的方向为沿着平行轴。对于六聚体来说,这一数值会小于0.1,这是由六聚体结构上不同Venus荧光蛋白质之间的局部homo FRET造成的。
单分子研究:通过反聚束探测单发射体
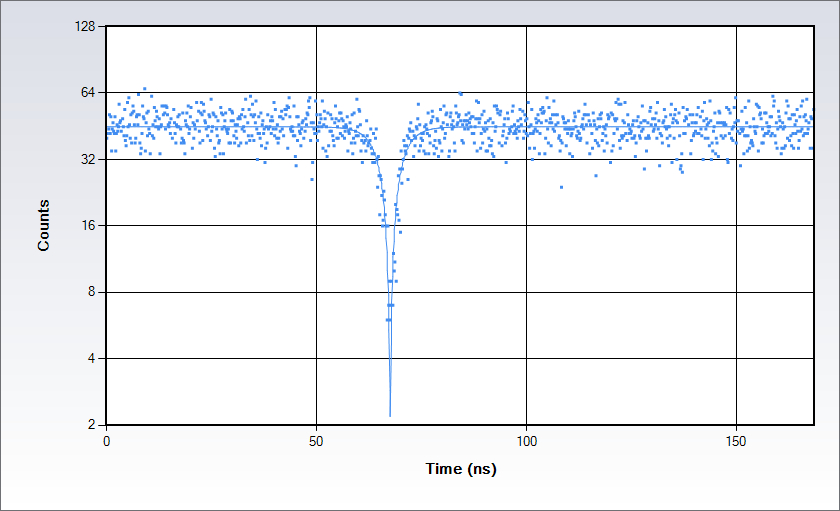
通过罗丹明110水溶液获得的反聚束数据 (488 nm处激发)。通过用经典Hanbury-Brown-Twiss装置将信号分成两束并把每个光束发送到Alba上分别的探测器以完成测量:采集电子设备能提供光子到达探测器处的时间差直方图。直方图在重合点处出现了下凹,凹陷的深度取决于观察体积中独立发射器的数量,而形状则由激发态寿命决定。
Product Specifications for Alba v5
仪器特点
- 每个采集通道上都有单独的针孔
- 通过电脑操控对针孔可变小孔的选择
- 通过电脑操控成像平面上针孔的位置
- 单光子或多光子激发
- 上至四个通道的数据采集
- 摄像头的辅助接口
显微镜
- 倒置和正置
物镜
- 拥有20X、40X、60X的放大倍率和1.5-8.1的工作距离
- 油浸物镜,1.4 NA和60X (标准);可提供其他小孔
- 水浸物镜,1.2 NA 60X (标准), 带有盖玻片校正 (用于0.15-0.18盖玻片);可以提供其他小孔
光源
- 单光子激光器被容纳在一个激光发射器内,通过电脑操控扩束器、激光强度和快门
- 多光子激发通过电脑操控扩束器、激光强度和快门
激光发射器
- 3、4、6激光器的型号。光通过单模式光纤被送至显微镜
振镜扫描仪
- 2个镀银扫描振镜
- 清晰的光学表面:3 mm
- 最大扫描率:5 KHz for 3 mm
- 扫描分辨率:64 x 64 to 4096 x 4096 pixels
- 扫描模式:Pt, Xt, XZ, XY, XZt, XYt, XYZ
- ROI扫描:长方形、椭圆、多边形、线
位置控制
- ISS 3轴控制单元
- ISS XY振镜控制单元
- ISS Z-压电控制单元
- 显微镜内置聚焦控制模块
- 自动化XY滑台 (ASI, Prior)
- XYZ压电台 (MadCity, PI)
针孔
- 孔径可变针孔;直径范围从20 µm至1000 µm
探测器
- 冷却GaAsP和GaAs PMT
- 冷却混合型PMT
- SPADs
二向色滤光片
- 用于单光子激发:1、2、3频段滤光片
- 用于多光子激发
偏振器
- 立方分光镜,波长范围:450 - 1100 nm; 消光比:在±3 度处为10,000:1
数据采集单元
- FastFLIM (数字频域FLIM)
- SWISS TCSPC卡(时域FLIM)
软件
- VistaVision
电脑和显示屏
- 高性能处理器, 32 GB RAM, Windows 11, 64比特
- 32"显示屏, 2556 x 1440分辨率
电源要求
- 110 - 240 V, 50/60 Hz, 400 VAC
大小 (mm)
- 885 (长) x 600 (宽) x 330 (高)
重量 (kg)
- 40 (不带显微镜)
Alba v5的测量
强度和寿命成像
- 1p或2p共焦图像
- 频域或TCSPC的FLIM
- 磷光寿命成像 (PLIM)
- 偏振图像
稳态和时间解析偏振各向异性成像
荧光波动光谱 (FFS)
- 荧光相关光谱 (FCS)
- 荧光互相关光谱 (FCCS)
- 光子计数直方图 (PCH)
- 荧光寿命相关光谱 (FLCS)
- 扫描FCS
- 数量和亮度 (N&B)
- 光栅成像相关光谱 (RICS)
粒子追踪 (自选的)
- 分子轨迹的3D重建
超分辨率
- 20 nm分辨率的纳米成像重建
单分子FRET突发分析
- 突发分析
- FRET和相关方法
- PIE-FRET方法
反聚束
Alba v5的产品配件
更多产品选择
-
灌注系统
这款蠕动泵为滑台上保持样本状态 (温度、pH等等) 提供了解决方案。
-
灌溉系统
当使用水物镜进行长时间测量时,它可以防止液体变干。
-
自动对焦保持
它通过使用主动反馈防止漂移,让物镜的焦点位置保持数个小时。
-
样本温度控制
滑台顶部的培养箱或是一个完整的外壳可以用于维持细胞培养的环境条件。
-
落射荧光灯
您可以通过Epi模块将您的样本可视化。请选择弧光灯或 LED 作为光源以及合适的滤光片立方体以添加到显微镜盒中。
-
原子力显微镜 (AFM)
与以下型号完全集成:
JPK-Bruker的NanoWizard
Bruker的Resolve
对于其他型号,请联系ISS。
Alba v5的产品软件
VistaVision
VistaVision是一款用于共焦显微镜应用的完整软件包,其中包括仪器控制、数据采集和数据处理。它易于使用,该软件易于使用,通过模块化组件开发,可在特定仪器配置被选择时激活。模块包括:
- FLIM/PLIM成像
- FFS
- smFRET
- 粒子追踪
产品资源
-
“Fluorescence lifetime imaging of physiological free Cu(ii) levels in live cells with a Cu(ii)-selective carbonic anhydrase-based biosensor.” Mccranor, B.J., Szmacinski, H., Zeng, H.H., Stoddard, A.K., Hurst, T., Fierke, C.A., Lakowicz, J.R. & Thompson, R.B. Metallomics, 6(5), p. 1034, 2014, Apr. doi: 10.1039/c3mt00305a.
-
“Application of Fluorescence Correlation Spectroscopy to Hapten–Antibody Binding.” Hazlett, T.L., Ruan, Q. & Tetin, S.Y. The Journal of Physical Chemistry B, 115(5), pp. 415–438, 2010, Dec. doi: 10.1385/1-59259-912-5:415.
-
“Determining Antibody Stoichiometry Using Time-Integrated Fluorescence Cumulant Analysis.” Skinner, J.P., Wu, B., Mueller, J.D. & Tetin, S.Y. The Journal of Physical Chemistry B, 115(5), pp. 1131–1138, 2010, Dec. doi: 10.1021/jp106279r.
-
“Fluorescence Correlation Spectroscopy Assay for Gliadin in Food.” Varriale, A., Rossi, M., Staiano, M., Terpetschnig, E., Barbieri, B., Rossi, M. & D'Auria, S. Analytical Chemistry, 79(12), pp. 4687–4689, 2007, May. doi: 10.1021/ac070475+.
-
“Antibodies in Diagnostic Applications.” Tetin, S. & Stroupe, S. Current Pharmaceutical Biotechnology, 5(1), pp. 9–16, 2004, Feb. doi: 10.2174/1389201043489602.
-
“Highly luminescent, biocompatible ytterbium(
iii ) complexes as near-infrared fluorophores for living cell imaging.” Ning, Y., Tang, J., Liu, Y.-W., Jing, J., Sun, Y. & Zhang, J.-L. Chemical Science, 9(15), pp. 3742–3753, 2018, May. doi: 10.1039/c8sc00259b. -
“Interleaflet Diffusion Coupling When Polymer Adsorbs onto One Sole Leaflet of a Supported Phospholipid Bilayer.” Zhang, L. & Granick, S. Macromolecules, 40(5), pp. 1366–1368, 2007, Feb. doi: 10.1021/ma062335s.
-
“How to Stabilize Phospholipid Liposomes (Using Nanoparticles).” Zhang, L. & Granick, S. Nano Letters, 6(4), pp. 694–698, 2006, Feb. doi: 10.1021/nl052455y.
-
“Slaved diffusion in phospholipid bilayers.” Zhang, L. & Granick, S. Proceedings of the National Academy of Sciences, 102(26), pp. 9118–9121, 2005, Jun. doi: 10.1073/pnas.0502723102.
-
“How Confined Lubricants Diffuse During Shear.” Mukhopadhyay, A., Bae, S.C., Zhao, J. & Granick, S. Physical Review Letters, 93(23), pp. 9118–9121, 2004, Dec. doi: 10.1103/physrevlett.93.236105.
-
“Polymer Lateral Diffusion at the Solid-Liquid Interface.” Zhao, J. & Granick, S. Journal of the American Chemical Society, 126(20), pp. 6242–6243, 2004, May. doi: 10.1021/ja0493749.
-
“Trapped Brownian Motion in Single- and Two-Photon Excitation Fluorescence Correlation Experiments.” Chirico, G., Fumagalli, C. & Baldini, G. The Journal of Physical Chemistry B, 106(10), pp. 2508–2519, 2002, Feb. doi: 10.1021/jp013087z.
-
“Stoichiometry of scaffold complexes in living neurons - DLC2 as a dimerization engine for GKAP.” Moutin, E., Compan, V., Raynaud, F., Clerté, C., Bouquier, N., Labesse, G., Ferguson, M.L., Fagni, L., Royer, C.A. & Perroy, J. Journal of Cell Science, 106(10), pp. 2508–2519, 2014, Jan. doi: 10.1242/jcs.145748.
-
“Reconciling molecular regulatory mechanisms with noise patterns of bacterial metabolic promoters in induced and repressed states.” Ferguson, M.L., Coq, D.L., Jules, M., Aymerich, S., Radulescu, O., Declerck, N. & Royer, C.A. Proceedings of the National Academy of Sciences, 109(1), pp. 155–160, 2011, Dec. doi: 10.1073/pnas.1110541108.
-
“Absolute quantification of gene expression in individual bacterial cells using two-photon fluctuation microscopy.” Ferguson, M.L., Coq, D.L., Jules, M., Aymerich, S., Declerck, N. & Royer, C.A. Analytical Biochemistry, 419(2), pp. 250–259, 2011, Dec. doi: 10.1016/j.ab.2011.08.017.
-
“Efficient Parallel Levenberg-Marquardt Model Fitting towards Real-Time Automated Parametric Imaging Microscopy.” Zhu, X. & Zhang, D. PLoS ONE, 8(10), p. e76665, 2013, Oct. doi: 10.1371/journal.pone.0076665.
-
“Nanometer-scale optical imaging of collagen fibers using gold nanoparticles.” Chen, B., Estrada, L.C., Hellriegel, C. & Gratton, E. Biomedical Optics Express, 2(3), p. 511, 2011, Feb. doi: 10.1364/boe.2.000511.
-
“Characterization of Brightness and Stoichiometry of Bright Particles by Flow-Fluorescence Fluctuation Spectroscopy.” Johnson, J., Chen, Y. & Mueller, J.D. Biophysical Journal, 99(9), pp. 3084–3092, 2010, Nov. doi: 10.1016/j.bpj.2010.08.057.
-
“Fluorescence correlation spectroscopy and photon counting histogram on membrane proteins: functional dynamics of the glycosylphosphatidylinositol-anchored urokinase plasminogen activator receptor.” Malengo, G., Andolfo, A., Sidenius, N., Gratton, E., Zamai, M. & Caiolfa, V.R. Journal of Biomedical Optics, 13(3), p. 031215, 2008, Nov. doi: 10.1117/1.2940570.
-
“Unraveling Protein-Protein Interactions in Living Cells with Fluorescence Fluctuation Brightness Analysis.” Chen, Y., Wei, L.-N. & Müller, J.D. Biophysical Journal, 88(6), pp. 4366–4377, 2005, Jun. doi: 10.1529/biophysj.105.059170.
-
“Dual-Color Photon-Counting Histogram.” Chen, Y., Tekmen, M., Hillesheim, L., Skinner, J., Wu, B. & Müller, J.D. Biophysical Journal, 88(3), pp. 2177–2192, 2005, Mar. doi: 10.1529/biophysj.104.048413.
-
“Fluorescence Spectroscopy with Metal–Dielectric Waveguides.” Badugu, R., Szmacinski, H., Ray, K., Descrovi, E., Ricciardi, S., Zhang, D., Chen, J., Huo, Y. & Lakowicz, J.R. The Journal of Physical Chemistry C, 119(28), pp. 16245–16255, 2015, Jul. doi: 10.1021/acs.jpcc.5b04204.
-
“Imaging of Protein Secretion from a Single Cell Using Plasmonic Substrates.” Szmacinski, H., Toshchakov, V., Piao, W. & Lakowicz, J.R. BioNanoScience, 3(1), pp. 30–36, 2013, Jan. doi: 10.1007/s12668-013-0076-7.
-
“Confocal Fluctuation Spectroscopy and Imaging.” Foldes-Papp, Z., Liao, S.-C.J., You, T., Terpetschnig, E. & Barbieri, B. Current Pharmaceutical Biotechnology, 11(6), pp. 639–653, 2010, Sep. doi: 10.2174/138920110792246618.
-
“Fluorescence fluctuation spectroscopy: ushering in a new age of enlightenment for cellular dynamics.” Jameson, D.M., Ross, J.A. & Albanesi, J.P. Biophysical Reviews, 1(3), pp. 105–118, 2009, Aug. doi: 10.1007/s12551-009-0013-8.
-
“Capturing directed molecular motion in the nuclear pore complex of live cells.” Cardarelli, F., Lanzano, L. & Gratton, E. Proceedings of the National Academy of Sciences, 109(25), pp. 9863–9868, 2012, Jun. doi: 10.1073/pnas.1200486109.
-
“Nanometer-scale imaging by the modulation tracking method.” Lanzano, L., Digman, M.A., Fwu, P., Giral, H., Levi, M. & Gratton, E. Journal of Biophotonics, 4(6), pp. 415–424, 2011, Apr. doi: 10.1002/jbio.201100002.
-
“A Zn-dependent structural transition of SOD1 modulates its ability to undergo phase separation.” Das, B., Roychowdhury, S., Mohanty, P., Rizuan, A., Chakraborty, J., Mittal, J. & Chattopadhyay, K. The EMBO Journal, 42(2), 2022, Nov. doi: 10.15252/embj.2022111185.
-
“Chromosome loading of cohesin depends on conserved residues in Scc3.” Pathania, A., Liu, W., Matityahu, A., Irudayaraj, J. & Onn, I. Current Genetics, 67(3), pp. 447–459, 2021, Jan. doi: 10.1007/s00294-020-01150-3.
-
“CAF-1 and Rtt101p function within the replication-coupled chromatin assembly network to promote H4 K16ac, preventing ectopic silencing.” Young, T.J., Cui, Y., Pfeffer, C., Hobbs, E., Liu, W., Irudayaraj, J. & Kirchmaier, A.L. PLOS Genetics, 16(12), p. e1009226, 2020, Dec. doi: 10.1371/journal.pgen.1009226.
-
“Copper mediates mitochondrial biogenesis in retinal pigment epithelial cells.” Dhivya, M.A., Aberami, S., Nikhalashree, S., Biswas, J., Liu, W., Irudayaraj, J., Sulochana, K., Coral, K. & Devi, S.B. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease, 1866(10), p. 165843, 2020, Oct. doi: 10.1016/j.bbadis.2020.165843.
-
“Quantitative High-Resolution Imaging of Live Microbial Cells at High Hydrostatic Pressure.” Bourges, A.C., Lazarev, A., Declerck, N., Rogers, K.L. & Royer, C.A. Biophysical Journal, 118(11), pp. 2670–2679, 2020, Jun. doi: 10.1016/j.bpj.2020.04.017.
-
“Monomeric cohesin state revealed by live-cell single-molecule spectroscopy.” Liu, W., Biton, E., Pathania, A., Matityahu, A., Irudayaraj, J. & Onn, I. EMBO reports, 21(2), 2019, Dec. doi: 10.15252/embr.201948211.
-
“Epigenetic biomarker screening by FLIM-FRET for combination therapy in ER+ breast cancer.” Liu, W., Cui, Y., Ren, W. & Irudayaraj, J. Clinical Epigenetics, 11(1), 2019, Jan. doi: 10.1186/s13148-019-0620-6.
-
“The N-Terminal Domain of ALS-Linked TDP-43 Assembles without Misfolding.” Tsoi, P.S., Choi, K.-J., Leonard, P.G., Sizovs, A., Moosa, M.M., Mackenzie, K.R., Ferreon, J.C. & Ferreon, A.C.M. Angewandte Chemie International Edition, 56(41), pp. 12590–12593, 2017, Sep. doi: 10.1002/anie.201706769.
-
“The Use of FLIM-FRET for the Detection of Mitochondria-Associated Protein Interactions.” Osterlund, E.J., Liu, Q. & Andrews, D.W. Angewandte Chemie International Edition, 56(41), pp. 395–419, 2015, Sep. doi: 10.1007/978-1-4939-2257-4_34.
-
“A Decoy Peptide that Disrupts TIRAP Recruitment to TLRs Is Protective in a Murine Model of Influenza.” Piao, W., Shirey, K., Ru, L., Lai, W., Szmacinski, H., Snyder, G., Sundberg, E., Lakowicz, J., Vogel, S. & Toshchakov, V. Cell Reports, 11(12), pp. 1941–1952, 2015, Jun. doi: 10.1016/j.celrep.2015.05.035.
-
“Measuring Förster Resonance Energy Transfer Using Fluorescence Lifetime Imaging Microscopy.” Day, R.N. Microscopy Today, 23(3), pp. 44–51, 2015, Apr. doi: 10.1017/s1551929515000395.
-
“Binding of fusion protein FLSC IgG1 to CCR5 is enhanced by CCR5 antagonist Maraviroc.” Latinovic, O., Schneider, K., Szmacinski, H., Lakowicz, J.R., Heredia, A. & Redfield, R.R. Antiviral Research, 112(5), pp. 80–90, 2014, Dec. doi: 10.1016/j.antiviral.2014.10.006.
-
“Cis and trans internucleosomal interactions of H3 and H4 tails in tetranucleosomes.” Nurse, N.P. & Yuan, C. Biopolymers, 103(1), pp. 33–40, 2014, Oct. doi: 10.1002/bip.22560.
-
“DNA Methylation Effects on Tetra-Nucleosome Compaction and Aggregation.” Jimenez-Useche, I., Nurse, N., Tian, Y., Kansara, B., Shim, D. & Yuan, C. Biophysical Journal, 107(7), pp. 1629–1636, 2014, Oct. doi: 10.1016/j.bpj.2014.05.055.
-
“Application of phasor plot and autofluorescence correction for study of heterogeneous cell population.” Szmacinski, H., Toshchakov, V. & Lakowicz, J.R. Journal of Biomedical Optics, 19(4), p. 046017, 2014, Apr. doi: 10.1117/1.jbo.19.4.046017.
-
“mMAPS: A Flow-Proteometric Technique to Analyze Protein-Protein Interactions in Individual Signaling Complexes.” Chou, C.-K., Lee, H.-H., Tsou, P.-H., Chen, C.-T., Hsu, J.-M., Yamaguchi, H., Wang, Y.-N., Lee, H.-J., Hsu, J.L., Lee, J.-F., Kameoka, J. & Hung, M.-C. Science Signaling, 7(315), pp. 517–524, 2014, Mar. doi: 10.1126/scisignal.2004620.
-
“Unexpected Complex Formation between Coralyne and Cyclic Diadenosine Monophosphate Providing a Simple Fluorescent Turn-on Assay to Detect This Bacterial Second Messenger.” Zhou, J., Sayre, D.A., Zheng, Y., Szmacinski, H. & Sintim, H.O. Analytical Chemistry, 86(5), pp. 2412–2420, 2014, Feb. doi: 10.1021/ac403203x.
-
“Unmethylated and methylated CpG dinucleotides distinctively regulate the physical properties of DNA.” Jimenez-Useche, I., Shim, D., Yu, J. & Yuan, C. Biopolymers, 101(5), pp. 517–524, 2014, Feb. doi: 10.1002/bip.22411.
-
“Unraveling transcription factor interactions with heterochromatin protein 1 using fluorescence lifetime imaging microscopy and fluorescence correlation spectroscopy.” Siegel, A.P., Hays, N.M. & Day, R.N. Journal of Biomedical Optics, 18(2), p. 025002, 2013, Feb. doi: 10.1117/1.jbo.18.2.025002.
-
“Monitoring Biosensor Activity in Living Cells with Fluorescence Lifetime Imaging Microscopy.” Hum, J., Siegel, A., Pavalko, F. & Day, R. International Journal of Molecular Sciences, 13(12), pp. 14385–14400, 2012, Nov. doi: 10.3390/ijms131114385.
-
“Chromophore maturation and fluorescence fluctuation spectroscopy of fluorescent proteins in a cell-free expression system.” Macdonald, P.J., Chen, Y. & Mueller, J.D. Analytical Biochemistry, 421(1), pp. 291–298, 2012, Feb. doi: 10.1016/j.ab.2011.10.040.
-
“HIV-1 Nef Binds a Subpopulation of MHC-I throughout Its Trafficking Itinerary and Down-regulates MHC-I by Perturbing Both Anterograde and Retrograde Trafficking.” Yi, L., Rosales, T., Rose, J.J., Chaudhury, B., Knutson, J.R. & Venkatesan, S. Journal of Biological Chemistry, 285(40), pp. 30884–30905, 2010, Oct. doi: 10.1074/jbc.m110.135947.
-
“Familial Hypertrophic Cardiomyopathy Can Be Characterized by a Specific Pattern of Orientation Fluctuations of Actin Molecules,.” Borejdo, J., Szczesna-Cordary, D., Muthu, P. & Calander, N. Biochemistry, 49(25), pp. 5269–5277, 2010, Jun. doi: 10.1021/bi1006749.
-
“Estrogen Receptor Interactions and Dynamics Monitored in Live Cells by Fluorescence Cross-Correlation Spectroscopy.” Savatier, J., Jalaguier, S., Ferguson, M.L., Cavaillès, V. & Royer, C.A. Biochemistry, 49(4), pp. 772–781, 2010, Jan. doi: 10.1021/bi9013006.
-
“Monomer-Dimer Equilibrium in Glutathione Transferases: A Critical Re-Examination.” Fabrini, R., Luca, A.D., Stella, L., Mei, G., Orioni, B., Ciccone, S., Federici, G., Bello, M.L. & Ricci, G. Biochemistry, 48(43), pp. 10473–10482, 2009, Oct. doi: 10.1021/bi901238t.
-
“A Fluorescent Mutant of the NM Domain of the Yeast Prion Sup35 Provides Insight into Fibril Formation and Stability.” Palhano, F.L., Rocha, C.B., Bernardino, A., Weissmuller, G., Masuda, C.A., Montero-Lomelí, M., Gomes, A.M., Chien, P., Fernandes, P.M.B. & Foguel, D. Biochemistry, 48(29), pp. 6811–6823, 2009, Jul. doi: 10.1021/bi9000276.
-
“Fluorescence Correlation Spectroscopy of Phosphatidylinositol-Specific Phospholipase C Monitors the Interplay of Substrate and Activator Lipid Binding.” Pu, M., Roberts, M.F. & Gershenson, A. Biochemistry, 48(29), pp. 6835–6845, 2009, Jul. doi: 10.1021/bi900633p.
-
“Correlation of Vesicle Binding and Phospholipid Dynamics with Phospholipase C Activity.” Pu, M., Fang, X., Redfield, A.G., Gershenson, A. & Roberts, M.F. Journal of Biological Chemistry, 284(24), pp. 16099–16107, 2009, Jun. doi: 10.1074/jbc.m809600200.
-
“Characterization of the Control Catabolite Protein of Gluconeogenic Genes Repressor by Fluorescence Cross-Correlation Spectroscopy and Other Biophysical Approaches.” Zorrilla, S., Ortega, Á., Chaix, D., Alfonso, C., Rivas, G., Aymerich, S., Lillo, M.P., Declerck, N. & Royer, C.A. Biophysical Journal, 95(9), pp. 4403–4415, 2008, Nov. doi: 10.1529/biophysj.108.135863.
-
“Endothelial adhesion receptors are recruited to adherent leukocytes by inclusion in preformed tetraspanin nanoplatforms.” Barreiro, O., Zamai, M., Yáñez-Mó, M., Tejera, E., López-Romero, P., Monk, P.N., Gratton, E., Caiolfa, V.R. & Sánchez-Madrid, F. Journal of Cell Biology, 183(3), pp. 527–542, 2008, Oct. doi: 10.1083/jcb.200805076.
-
“Applications of dual-color fluorescence cross-correlation spectroscopy in antibody binding studies.” Ruan, Q. & Tetin, S.Y. Analytical Biochemistry, 374(1), pp. 182–195, 2008, Mar. doi: 10.1016/j.ab.2007.11.007.
-
“Fructose-1,6-bisphosphate Acts Both as an Inducer and as a Structural Cofactor of the Central Glycolytic Genes Repressor (CggR).” Zorrilla, S., Chaix, D., Ortega, A., Alfonso, C., Doan, T., Margeat, E., Rivas, G., Aymerich, S., Declerck, N. & Royer, C.A. Biochemistry, 46(51), pp. 14996–15008, 2007, Dec. doi: 10.1021/bi701805e.
-
“Monomer–dimer dynamics and distribution of GPI-anchored uPAR are determined by cell surface protein assemblies.” Caiolfa, V.R., Zamai, M., Malengo, G., Andolfo, A., Madsen, C.D., Sutin, J., Digman, M.A., Gratton, E., Blasi, F. & Sidenius, N. Journal of Cell Biology, 179(5), pp. 1067–1082, 2007, Dec. doi: 10.1083/jcb.200702151.
-
“Escherichia coli ribosomal protein L20 binds as a single monomer to its own mRNA bearing two potential binding sites.” Allemand, F., Haentjens, J., Chiaruttini, C., Royer, C. & Springer, M. Nucleic Acids Research, 35(9), pp. 3016–3031, 2007, Apr. doi: 10.1093/nar/gkm197.
-
“Determining the stoichiometry of protein heterocomplexes in living cells with fluorescence fluctuation spectroscopy.” Chen, Y. & Müller, J.D. Proceedings of the National Academy of Sciences, 104(9), pp. 3147–3152, 2007, Feb. doi: 10.1073/pnas.0606557104.
-
“Interactions of Two Monoclonal Antibodies with BNP: High Resolution Epitope Mapping Using Fluorescence Correlation Spectroscopy.” Tetin, S.Y., Ruan, Q., Saldana, S.C., Pope, M.R., Chen, Y., Wu, H., Pinkus, M.S., Jiang, J. & Richardson, P.L. Biochemistry, 45(47), pp. 14155–14165, 2006, Nov. doi: 10.1021/bi0607047.
-
“Quantitative detection of the ligand-dependent interaction between the androgen receptor and the co-activator, Tif2, in live cells using two color, two photon fluorescence cross-correlation spectroscopy.” Rosales, T., Georget, V., Malide, D., Smirnov, A., Xu, J., Combs, C., Knutson, J.R., Nicolas, J.-C. & Royer, C.A. European Biophysics Journal, 36(2), pp. 153–161, 2006, Oct. doi: 10.1007/s00249-006-0095-1.
-
“Effects of Protein-Ligand Associations on the Subunit Interactions of Phosphofructokinase from B. stearothermophilus.” Quinlan, R.J. & Reinhart, G.D. Biochemistry, 45(38), pp. 11333–11341, 2006, Sep. doi: 10.1021/bi0608921.
-
“Phosphoinositide Specificity of and Mechanism of Lipid Domain Formation by Annexin A2-p11 Heterotetramer.” Gokhale, N.A., Abraham, A., Digman, M.A., Gratton, E. & Cho, W. Journal of Biological Chemistry, 280(52), pp. 42831–42840, 2005, Dec. doi: 10.1074/jbc.m508129200.
-
“The Nuclear Receptor Coactivator PGC-1α Exhibits Modes of Interaction with the Estrogen Receptor Distinct From those of SRC-1.” Bourdoncle, A., Labesse, G., Margueron, R., Castet, A., Cavaillès, V. & Royer, C.A. Journal of Molecular Biology, 347(5), pp. 921–934, 2005, Apr. doi: 10.1016/j.jmb.2005.01.048.
-
“Polarized fluorescence correlation spectroscopy of DNA-DAPI complexes.” Barcellona, M.L., Gammon, S., Hazlett, T., Digman, M.A. & Gratton, E. Microscopy Research and Technique, 65(4-5), pp. 205–217, 2004, Nov. doi: 10.1002/jemt.20121.
-
“Vesicle Encapsulation Studies Reveal that Single Molecule Ribozyme Heterogeneities Are Intrinsic.” Okumus, B., Wilson, T.J., Lilley, D.M. & Ha, T. Biophysical Journal, 87(4), pp. 2798–2806, 2004, Oct. doi: 10.1529/biophysj.104.045971.
-
“Pleckstrin Homology Domain Diffusion in Dictyostelium Cytoplasm Studied Using Fluorescence Correlation Spectroscopy.” Ruchira, , Hink, M.A., Bosgraaf, L., Haastert, P.J.v. & Visser, A.J. Journal of Biological Chemistry, 279(11), pp. 10013–10019, 2004, Mar. doi: 10.1074/jbc.m310039200.
-
“Probing protein oligomerization in living cells with fluorescence fluctuation spectroscopy.” Chen, Y., Wei, L.-N. & Müller, J.D. Proceedings of the National Academy of Sciences, 100(26), pp. 15492–15497, 2003, Dec. doi: 10.1073/pnas.2533045100.
-
“Segregation of Saturated Chain Lipids in Pulmonary Surfactant Filmsand Bilayers.” Nag, K., Pao, J.-S., Harbottle, R.R., Possmayer, F., Petersen, N.O. & Bagatolli, L.A. Biophysical Journal, 82(4), pp. 2041–2051, 2002, Apr. doi: 10.1016/s0006-3495(02)75552-5.
-
“Molecular Heterogeneity of O-Acetylserine Sulfhydrylase by Two-Photon Excited Fluorescence Fluctuation Spectroscopy.” Chirico, G., Bettati, S., Mozzarelli, A., Chen, Y., Müller, J.D. & Gratton, E. Biophysical Journal, 80(4), pp. 1973–1985, 2001, Apr. doi: 10.1016/s0006-3495(01)76167-x.
-
“Single-Molecule Tracking in Live Cell without Immobilization or without Hydrodynamic Flow by Simulations: Thermodynamic Jitter.” Baumann, G., and Földes-Papp, Z. Biophysica, 4, p. 442-452, 2024, Aug. doi: 10.3390/e22121380.
-
“Study on Single-molecule Biophysics and Biochemistry in Dilute Liquids and Live Cells without Immobilization or Significant Hydrodynamic Flow: The Thermodynamic Single-molecule Demon.” Foldes-Papp, Z. & Baumann, G. Current Pharmaceutical Biotechnology, 23(14), pp. 1750–1757, 2022, Nov. doi: 10.2174/1389201023666220616123928.
-
“Single-molecule time resolution in dilute liquids and live cells at the molecular scale: Constraints on the measurement time.” Földes-Papp, Z. American Journal of Translational Medicine, 5(3), pp. 154 - 165, 2021, Jan.
-
“Visualization of subdiffusive sites in a live single cell.” Földes-Papp, Z., Baumann, G. & Li, L.-C. Journal of Biological Methods, 8(1), p. e142, 2021, Jan. doi: 10.14440/jbm.2021.348.
-
“Measurements of Single Molecules in Solution and Live Cells Over Longer Observation Times Than Those Currently Possible: The Meaningful Time.” Foldes-Papp, Z. Current Pharmaceutical Biotechnology, 14(4), pp. 441–444, 2013, Apr. doi: 10.2174/1389201011314040009.
-
“Fluorescence Molecule Counting for Single-Molecule Studies in Crowded Environment of Living Cells without and with Broken Ergodicity.” Foldes-Papp, Z. & Baumann, G. Current Pharmaceutical Biotechnology, 12(5), pp. 824–833, 2011, May. doi: 10.2174/138920111795470949.
-
“Single actomyosin motor interactions in skeletal muscle.” Földes-Papp, Z., Liao, S.-C.J., Barbieri, B., Gryczynski, K., Luchowski, R., Gryczynski, Z., Gryczynski, I., Borejdo, J. & You, T. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, 1813(5), pp. 858–866, 2011, May. doi: 10.1016/j.bbamcr.2011.02.001.
-
“Meaningful Interpretation of Subdiffusive Measurements in Living Cells (Crowded Environment) by Fluorescence Fluctuation Microscopy.” Baumann, G., Place, R.F. & Foldes-Papp, Z. Current Pharmaceutical Biotechnology, 11(5), pp. 527–543, 2010, Aug. doi: 10.2174/138920110791591454.
-
“Anomalous behavior in length distributions of 3D random Brownian walks and measured photon count rates within observation volumes of single-molecule trajectories in fluorescence fluctuation microscopy.” Baumann, G., Gryczynski, I. & Földes-Papp, Z. Optics Express, 18(17), p. 17883, 2010, Aug. doi: 10.1364/oe.18.017883.
-
“Editorial [Hot Topic: Getting High on Single Molecule Biophysics (Guest Editor: Steven M. Block )].” Block, S. Current Pharmaceutical Biotechnology, 10(5), pp. 464–466, 2009, Aug. doi: 10.2174/138920109788922092.
-
“Reducing Background Contributions in Fluorescence Fluctuation Time- Traces for Single-Molecule Measurements in Solution.” Foldes-Papp, Z., Liao, S.-C., You, T. & Barbieri, B. Current Pharmaceutical Biotechnology, 10(5), pp. 532–542, 2009, Aug. doi: 10.2174/138920109788922137.
-
“Fluorescence Fluctuation Spectroscopic Approaches to the Study of a Single Molecule Diffusing in Solution and a Live Cell without Systemic Drift or Convection: A Theoretical Study.” Foldes-Papp, Z. Current Pharmaceutical Biotechnology, 8(5), pp. 261–273, 2007, Oct. doi: 10.2174/138920107782109930.
-
“Mobility and distribution of replication protein A in living cells using fluorescence correlation spectroscopy.” Braet, C., Stephan, H., Dobbie, I.M., Togashi, D.M., Ryder, A.G., Földes-Papp, Z., Lowndes, N. & Nasheuer, H.P. Experimental and Molecular Pathology, 82(2), pp. 156–162, 2007, Apr. doi: 10.1016/j.yexmp.2006.12.008.
-
“`True' single-molecule molecule observations by fluorescence correlation spectroscopy and two-color fluorescence cross-correlation spectroscopy.” Földes-Papp, Z. Experimental and Molecular Pathology, 82(2), pp. 147–155, 2007, Apr. doi: 10.1016/j.yexmp.2006.12.002.
-
“What it means to measure a single molecule in a solution by fluorescence fluctuation spectroscopy.” Földes-Papp, Z. Experimental and Molecular Pathology, 80(3), pp. 209–218, 2006, Jun. doi: 10.1016/j.yexmp.2006.01.001.
-
“How the Molecule Number Is Correctly Quantified in Two-Color Fluorescence Cross-Correlation Spectroscopy: Corrections for Cross-Talk and Quenching in Experiments.” Foldes-Papp, Z. Current Pharmaceutical Biotechnology, 6(6), pp. 437–444, 2005, Dec. doi: 10.2174/138920105775159296.
-
“Single-Phase Single-Molecule Fluorescence Correlation Spectroscopy (SPSM-FCS).” Földes-Papp, Z., Fuchs, J. & Podda, M. Encyclopedia of Medical Genomics and Proteomics, 1, pp. 1–7, 2004, Dec.
-
“Detection of single molecules: solution-phase single-molecule fluorescence correlation spectroscopy as an ultrasensitive, rapid and reliable system for immunological investigation.” Földes-Papp, Z., Demel, U. & Tilz, G.P. Journal of Immunological Methods, 260(1-2), pp. 117–124, 2002, Feb. doi: 10.1016/s0022-1759(01)00537-3.
Alba v5的配置
Alba可以选择两种几何配置中的一个:一个是分支配置,其中每一个通道荧光光束以级联形式分开;另一个是平行配置,其中荧光光束分别为通道1-2和3-4分离。

























